Systems Pharmacology and Molecular Docking Reveals the Potential Beneficial effect of Rabdosia serra in Treating liver Cancer
Article Information
Xinhui Niu1, †, Changsheng Dong2, †, Ismael Obaidi3, 4, Siying Chen5, Junying Liu*, 3
1 Department of Regenerative Medicine, School of Pharmaceutical Sciences, Jilin University, Changchun, China
2 Cancer Institute of Traditional Chinese Medicine, Longhua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
3 School of Pharmacy and Pharmaceutical Sciences, Trinity College Dublin, Dublin 2, Ireland
4 College of Pharmacy, University of Babylon, Babylon, Iraq
5The Second Affiliated Hospital, Nanchang University, Jiangxi, China
†XHN and CSD contributed equally to this work
*Corresponding author: Junying Liu. NatPro Center, School of Pharmacy and Pharmaceutical Sciences, Trinity College Dublin, Dublin 2, Ireland.
Received: 17 July 2023; Accepted: 26 July 2023; Published: 01 September 2023
Citation: Xinhui Niu, Changsheng Dong, Ismael Obaidi, Siying Chen, Junying Liu. Systems Pharmacology and Molecular Docking Reveals the Potential Beneficial effect of Rabdosia serra in Treating liver Cancer. Journal of Pharmacy and Pharmacology Research. 7 (2023): 145-156.
Share at FacebookAbstract
Introduction: The folk herb Rabdosia serra (Xi Huang Cao) has been used to treat several diseases, including jaundice, hepatitis, acute cholecystitis, and enteritis, for many years. Moreover, it is also one of the components of the Dan Shi Tong capsule, a medication that relieves the damp-heat symptoms in the gallbladder and liver. This research aimed to carry out a series of comprehensive pharmacological evaluations of the mechanism of action of this plant for the treatment of hepatocellular carcinoma (HCC).
Methods: This study determined the potential anticancer benefit of R. serra by predicting the molecular targets of the key components and intersecting the herbal targets with disease targets associated with liver cancer. The anti-hepatocellular carcinogenic value of R. serra and the potential influence on other liver disorders and malignancies were demonstrated using a systems pharmacology approach and molecular docking.
Results: Fishing for drug targets turned up 451 targets related to 20 important metabolites. A 216 intersected target set was produced by crossing drug targets (451) with HCC-associated elite targets (5725). These data were then used to analyse the protein-protein interaction (PPI) network and GO and KEGG pathways. A combination of these results revealed that the target factors (PIK3R1, RELA, EGFR, and EP300) were implicated in more than four signaling pathways, including the PI3K-Akt, mitogen-activated protein kinase (MAPK), Wnt, and p53 signaling routes. These findings were validated by molecular docking, which showed stable combinations between the five metabolites (linoleic acid, quercetin, caffeine, rutin, and methyl rosmarinate) and the four molecular targets (PIK3R1, RELA, EGFR, and EP300).
Conclusion: These findings establish the theoretical foundation for future study into the active drug-like components and mechanisms of action of R. serra in treating HCC and other hepatic disorders.
Keywords
Rabdosia serra; systems pharmacology; hepatocellular carcinoma; hepatoma; molecular docking
Rabdosia serra articles; systems pharmacology articles; hepatocellular carcinoma articles; hepatoma articles; molecular docking articles
Article Details
1. Introduction
Liver cancer (hepatoma) continues to be a global health challenge and a leading cause of cancer deaths worldwide, with a prediction of over 1 million cases by 2030 by the World Health Organization. Hepatocellular carcinoma (HCC) is the most common form of liver cancer, while hepatoblastoma mainly affects young children [1, 2]. HCC incidence remains a major oncologic challenge in many world areas, including Europe, the United States and Asian countries [3]. While liver transplant and resection can be curative in an early stage, it is often diagnosed at advanced stages of HCC, for which highly effective therapies are lacking [4, 5]. Recent years have seen a great diversification of treatment methods, such as lenvatinib, regorafenib, and sorafenib, approved as first or second-line treatment for advanced HCC patients [6]. Moreover, catheter-based intraarterial therapies have been demonstrated to be an attractive therapeutic option for patients who may have previously had other alternatives [7]. While we acknowledge that these are inspiring, their effectiveness is worth pointing out, which confers limited survival benefits.
Since irinotecan and topotecan, clinical derivatives of the plant alkaloid camptothecin, have been widely used as anticancer drugs for the past 20 years [8], the efforts to develop new medicines in oncology should continue focusing on natural products and the use of natural phytochemical compounds [9]. These natural therapies using plant-derived extracts may reduce adverse side effects compared with traditional cancer treatments [10]. In addition, the ability of phytochemicals to inhibit tumor formation by antioxidant, antiproliferative, and pro-apoptotic effects on a variety of conditions, including leukemia, prostate, breast, colon, brain, melanoma, and pancreatic cancers, has also been well documented [11]. However, the relative number of natural plant-based therapeutics, partially attributed to challenges associated with extraction, isolation, and their structural complexity, has steadily declined over the past several decades [12]. Thus, finding novel natural compounds through systems pharmacology based on big data simultaneously with low toxicity and high selectivity for killing cancer cells is an important area in cancer research [13].
Rabdosia serra (Maxim), a perennial plant, is found across South and Southeast Asia [16]. It is now demonstrated to have various pharmacological effects and has been used as a folk medicine for jaundice, hepatitis, acute cholecystitis and enteritis [17]. The bioactive molecules contained in the plant include diterpenoids (the most important active constituents contributing to the pharmacological efficacy), triterpenoids, flavonoids, phenolic acids and volatile oils [18]. Furthermore, R. serra, a valuable substance against jaundice and hypochondriac discomfort, treats damp heat in the liver and gallbladder in Traditional Chinese Medicine [19]. The top five chemicals tested in this investigation based on their degree values, linoleic acid, quercetin, caffeic acid, rutin, and methyl rosmarinate, were shown to interact with liver cancer-related molecular targets, which will be described in detail in the following sections in this paper.
Chinese medicine offers a wide range of potential avenues for investigation in regulating liver function and modulating abnormalities in numerous organ systems. Recently, research has focused on novel drug discovery for treating liver cancers via systems pharmacology [20]. As a result, we believe that elucidating the structure and molecular targets of the complex components of herbal plants and discovering or repurposing existing medications will be an alternate development avenue for cancer treatment.
2. Methods and Materials
2.1 Compound Screening and Targeted Collection
The compounds needed for this project were obtained by literature matching and collecting the main active metabolites derived from R. serra based on China Knowledge Network (CNKI: https://www.cnki.net/), Bioinformatics Analysis Tool for Molecular Mechanisms in Chinese Medicine (BATMAN-TCM: http://bionet.ncpsb.org.cn/batman-tcm/index.php), Encyclopedia of Traditional Chinese Medicine (ETCM: http://www.tcmip.cn/ETCM/index.php/Home/Index/) and High-Entomology Database of Traditional Chinese Medicine (HERB: http://herb.ac.cn/) [19, 21]. Subsequently, the SMILEs and other relevant information were gained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). The names of the molecules and their chemical structure formulas were checked, and the oral bioavailability (OB) and drug-like (DL) information of the flavonoids were retrieved from TCMSP (http: //tcmspw.com /tcmsp.php) to establish a database of flavonoids [22]. The Similarity Ensemble Approach (SEA: https://sea.bkslab.org/) with target species setting as Homo sapiens was then used to remove duplications were removed; the BATMAN-TCM (score >15, P<0.05) and HERB databases were applied to find relevant targets for the compounds [21, 23].
2.2 Acquisition of Intersectional Targets between the Components of Rabdosia serra and HCC
The DisGeNET database (https://www.disgenet.org/search) was searched using "Liver carcinoma" to obtain relevant targets for liver cancer disease. The Bioinformatics & Evolutionary Genomics website was subsequently used to intersect targets with liver cancer targets and get a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) [24].
2.3 Construction of Protein-Protein Interaction Network
The intersecting targets were imported into the STRING database under the "multiple proteins" option, the species of origin was selected as Homo sapiens for "organism," targets with the highest confidence score (>0.9) were set, and the free nodes were hidden [25]. The protein-protein interaction (PPI) network analysis of the effect of the components of R. serra on HCC was carried out. The higher the value, the more central the target is in the PPI network. The ".tsv" file was downloaded from the STRING database that generates information about the PPI network, which was further imported into Cytoscape 3.6.0. The cytoHubba function was chosen to filter the core targets.
2.4 GO and KEGG Enrichment Analysis
The functional annotation, classification, and enrichment analysis of common targets were performed using Metascape and the organism was limited to Homo sapiens [26]. The key cancer gene pathways in Metascape were then shown using KEGG mapper, along with therapeutic targets for each of these pathways [27-29]. Further analysis of the genes was performed through the DAVID platform [30, 31].
2.5 Component-Target-Pathway Network Construction
The components, intersecting targets and major pathways of R. serra were imported into Cytoscape 3.6.0 software to construct a component-target-pathway network map of R. serra components against liver cancer. The topological analysis was performed using the Network Analyzer function, and the top five components were selected for molecular docking validation based on their degree values.
2.6 Molecular Docking
The 2D molecular structures of the ligands and their active components were obtained from PubChem, and the crystal structures of the primary target proteins were acquired from RCSB PDB (https://www.wwpdb.org/) to verify the binding of the hub genes to their corresponding components. Autodock Vina 1.5.6 was used for molecular docking. The molecule with the lowest binding energy in the docked conformation was chosen; the lower the binding energy, the simpler it is for the active component to attach to the receptor. The molecular docking pattern is finally visualized by PyMOL 2.3.4.
3. Results
3.1 Target Prediction and Analysis
We found 23 components based on the literature review: BATMAN-TCM, HERB, and ETCM. In addition, the TCMSP database was searched, and only five components with oral bioavailability (OB) ≥ 20% were finally identified, while only three components with drug-likeness (DL) ≥ 0.1 were identified. Further, for the targets of the obtained compounds, a search of the potential targets based on the BATMAN-TCM (score >15, P<0.05), SEA and HERB databases resulted in a total of 451 targets associated with 20 components. In contrast, the OB and DL values of 12 components obtained in the TCMSP database are shown in Table 1.
Table 1: Information of 12 flavonoid compounds from TCMSP
|
Components |
CAS Number |
Oral bioavailability, OB (%) |
Drug-like, DL (%) |
|
Caffeic acid |
331-39-5 |
54.97 |
0.05 |
|
Isorhamnetin |
480-19-3 |
49.6 |
0.31 |
|
Quercetin |
117-39-5 |
46.43 |
0.28 |
|
Loliolide |
06-02-5989 |
44.66 |
0.08 |
|
Linoleic acid |
6144-28-1 |
41.9 |
0.14 |
|
Corosolic Acid |
4547-24-4 |
18.56 |
0.74 |
|
Ursolic acid |
77-52-1 |
16.77 |
0.75 |
|
Totarol |
511-15-9 |
15.78 |
0.25 |
|
Chlorogenic acid |
327-97-9 |
11.93 |
0.33 |
|
Rutin |
153-18-4 |
3.2 |
0.68 |
|
Rosmarinic acid |
20283-92-5 |
1.38 |
0.35 |
|
Methyl rosmarinate |
99353-00-1 |
1.37 |
0.37 |
3.2 Collection of Intersectional Targets
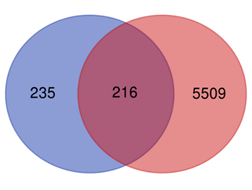
The dataset on liver cancer genes was retrieved from the DisGeNET database and paired with the constituent target ensemble we previously obtained, yielding 216 overlapping targets as prospective targets of R. serra constituents for the therapy of liver cancer using Venn diagram mapping (Figure 1).
3.3 Network Analysis and Construction
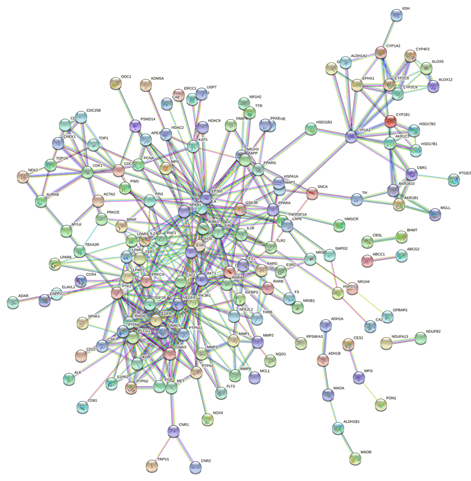
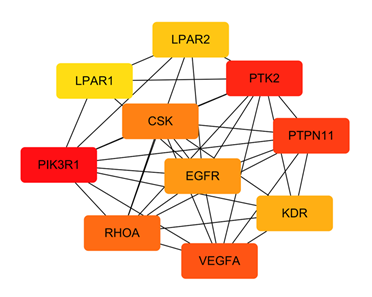
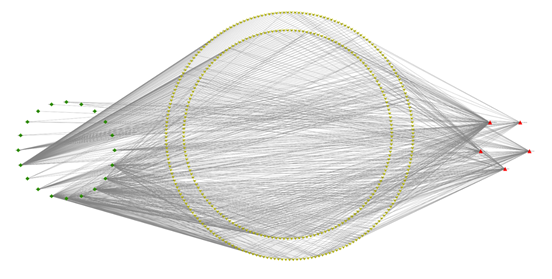
The STRING database was used to import the 216 intersection targets, and the disconnected nodes were removed to create a PPI network with a confidence level > 0.9. Figure 2 depicts the PPI network, which has 221 nodes and 438 edges with an average node degree value of 3.96. We next imported the network into Cytoscape 3.6.0 and utilised the cytoHubba plugin to identify the hub gene, as shown in Figure 3. The top ten genes calculated by the MCC algorithm were listed according to the scores: PIK3R1, PTK2, PTPN11, VEGFA, RHOA, CSK, EGFR, KDR, LPAR2, and LPAR1 (Table 3). The MNC, EPC, Degree, and Closeness algorithms were applied to determine the top 10 hub targets, and the results are displayed in Table 2. The targets in the table in black italics are important, and we chose the top objectives as docking targets based on the number of repetitions and the order of ranking (PIK3R1, RELA, EGFR and EP300).
216 overlapping targets as prospective targets of R. serra constituents for the therapy of liver cancer using Venn diagram mapping (Figure 1).Table 2: Top 10 targets in PPI network ranked by MCC method.
|
Rank |
Name |
Score |
|
1 |
PIK3R1 |
2009 |
|
2 |
PTK2 |
1801 |
|
3 |
PTPN11 |
1796 |
|
4 |
VEGFA |
1732 |
|
5 |
RHOA |
1692 |
|
6 |
CSK |
1440 |
|
7 |
EGFR |
1268 |
|
8 |
KDR |
786 |
|
9 |
LPAR2 |
510 |
|
10 |
LPAR1 |
499 |
Table 3: Different calculation methods and key genes
|
Category |
Rank methods in CytoHubba |
||||
|
EPC |
MCC |
MNC |
Degree |
Closeness |
|
|
1 |
PIK3R1 |
PIK3R1 |
PIK3R1 |
EP300 |
EP300 |
|
2 |
RELA |
PTK2 |
EP300 |
PIK3R1 |
RELA |
|
3 |
EGFR |
PTPN11 |
RELA |
RELA |
PIK3R1 |
|
4 |
EP300 |
VEGFA |
AKT1 |
AKT1 |
JUN |
|
5 |
AKT1 |
RHOA |
NFKB1 |
EGFR |
AKT1 |
|
6 |
PTK2 |
CSK |
ESR1 |
NFKB1 |
NFKB1 |
|
7 |
RHOA |
EGFR |
JUN |
ESR1 |
ESR1 |
|
8 |
VEGFA |
KDR |
EGFR |
VEGFA |
VEGFA |
|
9 |
ESR1 |
LPAR2 |
PTPN11 |
JUN |
EGFR |
|
10 |
PTPN11 |
LPAR1 |
RHOA |
PTPN11 |
PRKCA |
3.4 Functional Classification and Enrichment Analysis of the Common Gene Sets
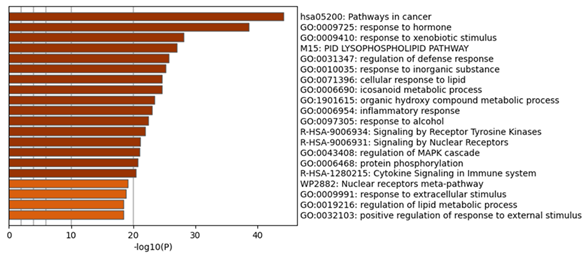
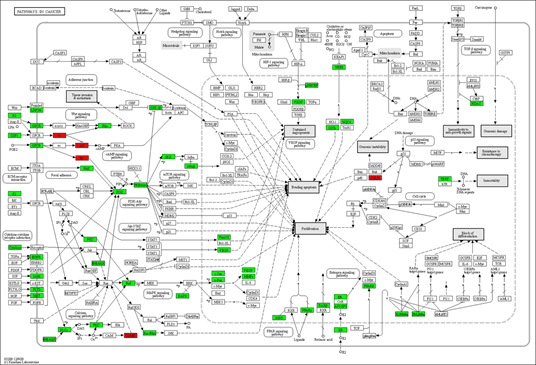
In the process and the KEGG/Wiki/Canonical route, the common R. serra-HCC targets were significantly enriched, according to the Metascape study (Figure 4). The top 20 enrichment terms by the p-value are pathways in cancer (hsa05200), response to hormone (GO:0009725), response to xenobiotic stimulus (GO:0009410), lysophospholipid pathway (M15-PID), regulation of defense response (GO:0031347), response to inorganic substance (GO:0010035), cellular response to lipid (GO:0071396), icosanoid metabolic process (GO:0006690), organic hydroxy compound metabolic process (GO:1901615), inflammatory response (GO:0006954), response to alcohol (GO:0097305), signaling by receptor tyrosine kinases (R-HSA-9006934), signaling by nuclear receptors (R-HSA-9006931), regulation of the mitogen-activated protein kinase (MAPK) cascade (GO:0043408), protein phosphorylation (GO:0006468), cytokine signaling in immune system (R-HSA-1280215), nuclear receptors meta-pathway (WP2882), response to extracellular stimulus (GO:0009991), regulation of lipid metabolic process (GO:0019216), positive regulation of response to external stimulation (GO:0032103). Of these, the cancer pathway was the most significantly enriched.
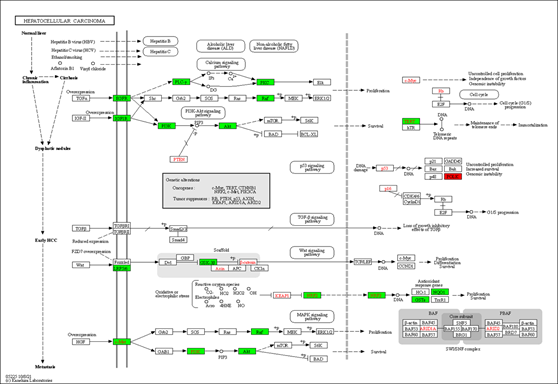
We also used the KEGG mapper to visualize the herb's additional targets enriched for the cancer pathway (hsa05200, Figure 5). The typical herbal cancer targets are shown in green, whereas other herbal targets are highlighted in red. As shown in Figure 5, the targets EGRF, MET, Raf, and DAPK are associated with the MAPK signaling pathway, while the herb's major targets are also the PI3K-Akt route. There are also many drug targets in the Wnt and the JAK-STAT signaling pathway. Similarly, in the pathway analysis of HCC (Figure 6), GSK-3 controlled cell proliferation, differentiation, and survival, whereas TERT influenced telomerase activity and KEAP1-NRF2-associated targets influenced cell survival. Finally, one of the downstream targets associated with TP53 transcription was found to be the POLK target. Further, the DAVID platform was used to analyze the genes and select the "KEGG_PATHWAY" and "WIKIPATHWAYS" options. As a result, 22 targets were involved in the PI3K-Akt signaling pathway (p<0.05), and 18 were involved in the PI3K-Akt-mTOR (the mechanistic target of rapamycin)-signaling pathway (p<0.05). The "DISGENET" and "OMIM_DISEASE" options were further selected, and the higher Classification Stringency condition showed that this ensemble was also associated with Breast Carcinoma (p<0.05, 26 targets), Adenocarcinoma (p <0.05, 10 targets) and Diffuse Astrocytoma (p<0.05, 6 targets).
3.5 Component-Target-Pathway Network Construction
A network of 20 flavonoid components, 216 crossing targets, and five significant pathways was created using Cytoscape 3.6.0 (Figure 6). There are 404 nodes and 1026 edges in the network diagram, with 20 green diamond nodes representing components of R. serra, 216 red inverted triangles representing intersection targets, five red triangles representing critical pathways, and the edges representing the relationship between components, targets, and pathways (Figure 7). To validate molecular docking, the top five components were chosen based on their degree values: rutin (degree:82), caffeic acid (degree:97), quercetin (degree:97), linoleic acid (degree:152), and methyl rosmarinate (degree:60).
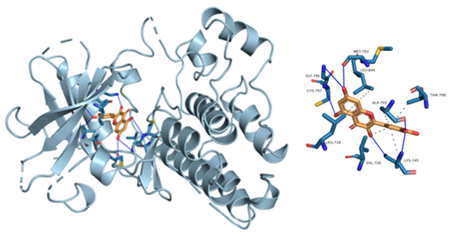
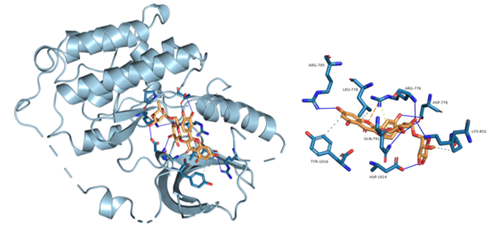
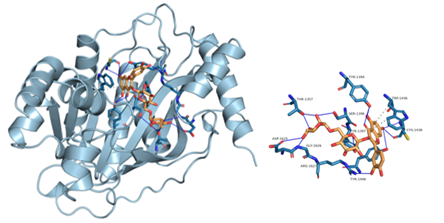
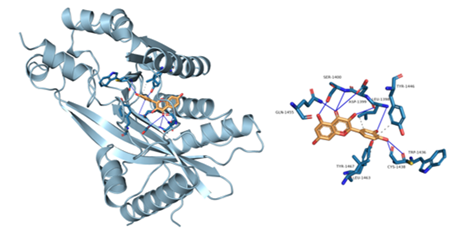
Figure 8: Component-target molecular docking interaction. The binding affinities are presented in Table 4.
3.6 Molecular Docking
Lower energy and a higher chance of contact resulted from a more stable ligand conformation when it binds to the receptor. The foundation for calculating interaction capacity is the binding energy of -5 kJ/mol. Linoleic acid (CAS: 6144-28-1), quercetin (CAS: 117-39-5), caffeic acid (CAS: 331-39-5), rutin (CAS: 153-18-4) and methyl rosmarinate (CAS: 99353-00-1) were selected as small molecule ligands, and EGFR (PDB ID: 5UGB), PIK3R1(PDB ID: 5LKZ), EP300 (PDB ID: 1H9O) and RELA (PDB ID: 3RCO) were molecularly docked as protein receptors (Figure 8). All five components' binding energies were negative, as shown in Table 4, demonstrating their capacity to attach to the target site spontaneously. However, all five parts showed binding solid affinities to the EGFR and PIK3R1. On the other hand, linoleic acid has a poor capacity to interact with EP300 and RELA. Based on the whole picture, molecular docking revealed that the chytrid components exhibit superior binding potential to the key targets.
Table 4: Molecular docking results of some components with targets
|
Target name |
PDB ID |
Ingredient |
Binding affinity (kcal/mol) |
|
EGFR |
5UGB |
Caffeic acid |
-6.2 |
|
Linoleic acid |
-5.1 |
||
|
Methyl rosmarinate |
-5.9 |
||
|
Quercetin |
-8.9 |
||
|
Rutin |
-8 |
||
|
EP300 |
5LKZ |
Caffeic acid |
-6.5 |
|
Linoleic acid |
-4.1 |
||
|
Methyl rosmarinate |
-6.9 |
||
|
Quercetin |
-8.9 |
||
|
Rutin |
-8.8 |
||
|
PIK3R1 |
1H9O |
Caffeic acid |
-5.8 |
|
Linoleic acid |
-5.3 |
||
|
Methyl rosmarinate |
-6.7 |
||
|
Quercetin |
-7.1 |
||
|
Rutin |
-6.3 |
||
|
RELA |
3RCO |
Caffeic acid |
-5.1 |
|
Linoleic acid |
-3.8 |
||
|
Methyl rosmarinate |
-5.6 |
||
|
Quercetin |
-6.4 |
||
|
Rutin |
-7.1 |
4. Discussion
Hepatocellular carcinoma is the most prevalent type of primary liver cancer [32]. It occurs most commonly in chronic liver disease, typically brought on by infection with hepatitis B or hepatitis C and regular alcohol consumption [33]. Although surgery is now the most popular treatment option for HCC, the traits of multifocal growth and distant metastases prevent surgical treatment from being curative in most HCC patients [34]. In addition, the five-year survival rate for HCC patients is still poor due to metastasis, dissemination, and high recurrence rates [35]. Therefore, it is anticipated that systems pharmacology analysis will enable us to reevaluate the effectiveness value of the existing available medications or herbal remedies, enhancing cancer patients' outcomes and experiences.
In this research, we analyzed the components of R. serra. The top five components (linoleic acid, quercetin, caffeic acid, rutin, and methyl rosmarinate) were filtered based on the degree value from the network diagram created. Linoleic acid, an important unsaturated fatty acid, has an isomer called conjugated linoleic acid (CLA) [36]. Conjugated linoleic acid (CLA; cis-9, trans-11-18:2), one of the most desired unsaturated fatty acids with the healthiest features, is the unsaturated fatty acid [37]. Natural flavonoid quercetin has been a promising therapeutic ligand for human triple-negative breast cancer, which inhibits various target proteins, including heat shock factor 1 (HSF1), Nuclear factor kappa B (NF-κB), CYP3A4 and numerous kinases [38]. Inhibition of HSF1 can make the antiproliferative effects of simvastatin in HCC cells more noticeable since HSF1 supports the growth and progression of malignancy [39]. It has also been demonstrated that HSF1 directly triggers miR-135b expression, promoting HCC cell motility and invasiveness, whereas miR-135b increases HCC cell metastasis in vivo and migration and invasion in vitro [40].
Similarly, NF-κB is constitutively active in some malignancies controlling genes involved in cell survival and proliferation, and it is also a key regulator of inflammatory responses [41]. It has also been shown that NF-κB plays a crucial role in modulating the development of liver disorders and even cancer by activating survival and inflammation of damaged hepatocytes, inducing spontaneous liver injury, fibrosis, and HCC [42]. About half of all commercially available medications are metabolized by CYP3A4, an important CYP450 enzyme responsible for the oxidation and inactivation of the anticancer agent taxol [43]. Caffeic acid and its derivative, caffeic acid phenethyl ester CAPE, have shown therapeutic benefits of reducing multidrug resistance in cancer cells by locking the efflux function of human P-glycoprotein against lung and breast cancer cells [44]. Additionally, the C/EBP Homologous Protein (CHOP)-mediated apoptosis was induced in hepatocarcinoma when Hep3B cells were treated with caffeic acid phenethyl ester, and DR5 protein levels were elevated [45]. Rutin was non-toxic to hepatoma cells, dramatically blocking the binding between HCV-LP and hepatoma cells and preventing cell-culture-derived HCV (HCVcc) from entering hepatoma cells [46]. It has been reported as a hepatoprotective, antioxidant, and anti-inflammatory agent [47]. Among Rosmarinus acid derivatives, methyl rosmarinate provided notable antioxidant and AR-inhibiting properties [48]. As a result, the above-mentioned key components of R. serra may have a role in intervening in liver cancer by regulating cell proliferation and apoptosis, reducing oxidative stress, decreasing drug resistance build-up, and down-regulating inflammatory levels.
According to the core target network screening, PIK3R1, RELA, EGFR, EP300, and other core targets are among the targets of R. serra components that affect liver cancer. First of all, aberrant PI3K signaling is a hallmark of cancer. As one of the key players in the PI3K pathway, which is linked to tumor growth and metastasis, PIK3R1 can control cancer cell proliferation [49]. In HCC clinical samples, PIK3R1 expression was elevated, and its silencing decreased the expression of p-PI3K, p-AKT, and p-mTOR, limiting cell proliferation, migrating, and expediting apoptosis in HCC cells [50]. It has also been demonstrated that mTOR signaling increases the production of new fatty acids [51]. As a potential target for cancer therapy, PIK3R1, therefore, deserves further exploration. Additionally, RELA, a protein-coding gene known as p65, is a widely expressed pleiotropic NF-kB subunit that stimulates the expression of the target gene IRF1 when RELA levels and baseline transcriptional activity are increased [52]. Furthermore, overexpression of RELA/p65 inhibits the growth of breast cancer cells by activating IRF1 in a manner that relies on RELA/p65 [53]. The RELA/p65 binding site in the Fas promoter was previously found and it has been demonstrated that tumor necrosis factor (TNF)-induced NF-κB-mediated elevation of Fas also further mediates the apoptosis [54]. Thus, regulating the expression of the RELA gene and related products is likely a potential therapeutic approach. Overexpression of the epidermal growth factor receptor (EGFR) and EGFR-activating mutations have been documented in numerous human tumors and are linked with poor clinical outcomes [55]. Additionally, EGFR promotes tumor stem/progenitor cell expansion and survival, supporting the use of EGFR-targeted drugs in managing resistant malignancies [56]. According to the abnormal Wnt signaling connected to HCC patients, proteins are usually regarded as the prominent oncogenes in HCC. They promote the signaling of growth factor receptors like EGFR to promote the survival of tumor cells [57]. As a histone acetyltransferase, EP300 plays a key role in regulating transcriptional activity through chromatin remodeling, thereby influencing cell proliferation and differentiation [58]. The mTOR complex 1 (mTORC1) is a key cell development and metabolism regulator. Acetyl-coenzyme A (AcCoA), a Leu metabolite, positively controls mTORC1 activity by acetylating raptor, an important mTORC1 regulator, at K1097 [59]. In conclusion, it is unequivocal that the anticipated action targets of R. serra are implicated in the etiology of HCC. As a result, we propose that the R. serra constituents may possess potential therapeutic benefits by acting on these critical targets.
Cirrhosis and chronic hepatitis account for over 90% of HCC cases [60]. The findings of the combined enrichment indicate that the therapies for treating HCC associated with R. serra components could have anticancer effects through essential pathways such as the PI3K/Akt, MAPK, Wnt, calcium, and p53 signaling pathways. The balance between cell survival and apoptosis is maintained through the PI3K/PTEN/AKT/mTOR pathway, which also plays a significant part in the emergence of resistance to platinum compounds, taxanes, and fluoropyrimidines [61]. The kinase cascade that results from the activation of PI3K by EGFR, VEGFR, and PDGFR can further provide signals that promote cell growth, survival, and antigenicity through AKT and mTOR [61, 62]. It has been demonstrated that the PI3K-AKT-mTOR signaling route is necessary for the survival and proliferation of luminal breast cancers, making them highly vulnerable to the blockage of pathway elements [63]. This concurs with our findings on adenocarcinoma and breast cancer. The most common survival signals that encourage the growth of hepatocellular carcinoma are NF-κB, Akt, MAPK, and mTOR, and intricate connections between the mTOR and MAPK pathways have been shown in hepatocarcinogenesis [64]. The evolution of liver fibrosis is aided by the PI3K/Akt pathway, which controls HCC activation, cell proliferation, and collagen production. Liver fibrosis can proceed to cirrhosis, which causes organ failure and death [65]. Hepatitis, hepatic steatosis, hepatic fibrosis, as well as infection with the hepatitis B and C viruses are the primary risk factors for the development of HCC [66]. The Wnt pathway is closely linked to an oncogenic phenotype in many malignancies, including liver, breast, and colon. The Wnt ligand receptor FZD7 is implicated in the initiation and progression of HCC, and increased expression of NF-kB-related Wnt-1 may be a key mechanism in hepatocarcinogenesis. Hepatocarcinogenesis is usually linked with aberrant activation of the Wnt/-catenin pathway [67]. As a transcription factor, p53 responds to stress signals and controls the expression of its target genes, causing a range of cellular responses to prevent tumor formation. P53 is the most mutated gene in human malignancies [68]. Due to the composition complexity and the target diversity, herbal compounding provides a promising field of research in drug discovery and development when a class of medications ameliorates the pathology of a type of organ.
Acknowledgments
We want to thank the financial support of H2020 Marie Sklodowska-Curie Actions and Enterprise Ireland (No.713654).
Funding
H2020 Marie Sklodowska-Curie Actions and Enterprise Ireland (No.713654), Shanghai Municipal Health Commission (No. 202040155), and Shanghai Municipal Science and Technology Commissions Special Biomedical Technology Support Plan (No. 20S31904100).
Statement of Conflict
No conflict of interest
References
- Lindblad KE, Ruiz de Galarreta M, Lujambio A. Tumor-intrinsic mechanisms regulating immune exclusion in liver cancers. Front Immunol 12 (2021): 642958.
- Schaller E, Ma A, Gosch LC, Klefenz A, Schaller D, Goehringer N, et al. New 3-Aryl-2-(2-thienyl) acrylonitriles with high activity against hepatoma cells. Int J Mol Sci 22 (2021).
- Chapiro J, Duran R, Lin M, Schernthaner RE, Wang Z, Gorodetski B, et al. Identifying staging markers for hepatocellular carcinoma before transarterial chemoembolization: comparison of three-dimensional quantitative versus non-three-dimensional imaging markers. Radiology 275 (2015): 438-447.
- El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389 (2017): 2492-2502.
- Waghray, A, Murali, A.R, Menon, K.N. Hepatocellular carcinoma: From diagnosis to treatment. World J Hepatol 7 (2015): 1020-1029.
- Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, et al. β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov 9 (2019): 1124-1141.
- Lewandowski RJ, Geschwind JF, Liapi E, Salem R. Transcatheter intraarterial therapies: rationale and overview. Radiology 259 (2011): 641-657.
- Thomas A, Pommier Y. Targeting topoisomerase I in the era of precision medicine. Clin Cancer Res 25 (2019): 6581-6589.
- Suttana W, Mankhetkorn S, Poompimon W, Palagani A, Zhokhov S, Gerlo S, et al. Differential chemosensitization of P-glycoprotein overexpressing K562/Adr cells by withaferin A and Siamois polyphenols. Mol Cancer 9 (2010): 99.
- Yamamoto T, Uemura K, Moriyama K, Mitamura K, Taga A. Inhibitory effect of maple syrup on the cell growth and invasion of human colorectal cancer cells. Oncol Rep 33 (2015): 1579-1584.
- Kawasaki BT, Hurt EM, Mistree T, Farrar WL. Targeting cancer stem cells with phytochemicals. Mol Interv 8 (2008): 174-184.
- Parker CG, Kuttruff CA, Galmozzi A, Jørgensen L, Yeh CH, Hermanson DJ, et al. Chemical proteomics identifies SLC25A20 as a functional target of the ingenol class of actinic keratosis drugs. ACS Cent Sci 3 (2017): 1276-1285.
- Wang S, Liu Q, Zhang Y, Liu K, Yu P, Liu K, et al. Suppression of growth, migration and invasion of highly metastatic human breast cancer cells by berbamine and its molecular mechanisms of action. Mol Cancer 8 (2009): 81.
- Jin H, Lu L, Liu J, Cui M. COVID-19 emergencies around the globe: China's experience in controlling COVID-19 and lessons learned. Int J Qual Health Care 33 (2021).
- Tong T, Wu YQ, Ni WJ, Shen AZ, Liu S. The potential insights of Traditional Chinese Medicine on treatment of COVID-19. Chin Med 15 (2020): 51.
- China TPCoPsRo. Pharmacopoeia of the People's Republic of China; Chinese Medical Science and Technology Press: Beijing (2010): 26.
- Wan J, Jiang H-Y, Tang J-W, Li X-R, Du X, Li Y, et al. Ent-abietanoids isolated from Isodon serra. Molecul 22 (2017).
- Xie W, Wen X, Zhang D, Zhang Y, Zhang Z, Jin Y, et al. Network pharmacology-based strategy to investigate harmacological mechanisms of Isodon serra (Maxim.) Hara for treatment of inflammatory. J Chinese Pharm Sci 31 (2022): 14.
- Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res 47 (2019): D976-d982.
- Li Z, Li Q, Wu J, Wang M, Yu J. Artemisinin and Its Derivatives as a Repurposing Anticancer Agent: What Else Do We Need to Do? Molecules 21 (2016).
- Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep 6 (2016): 21146.
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics 6 (2014): 13.
- Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol 25 (2007): 197-206.
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research 48 (2020): D845-D855.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47 (2019): D607-d613.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10 (2019): 1523.
- Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Science 28 (2019): 1947-1951.
- Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Research 49 (2021): D545-D551.
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28 (2000): 27-30.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 (2009): 44-57.
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 50 (2022): W216-221.
- Tseng HC, Xiong W, Badeti S, Yang Y, Ma M, Liu T, et al. Efficacy of anti-CD147 chimeric antigen receptors targeting hepatocellular carcinoma. Nat Commun 11 (2020): 4810.
- Yan G, Wang X, Sun C, Zheng X, Wei H, Tian Z, et al. Chronic alcohol consumption promotes Diethylnitrosamine-induced hepatocarcinogenesis via immune disturbances. Sci Rep 7 (2017): 2567.
- Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer 18 (2019): 105.
- Huang JL, Cao SW, Ou QS, Yang B, Zheng SH, Tang J, et al. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer 17 (2018): 93.
- Fujita Y, Kano K, Kishino S, Nagao T, Shen X, Sato C, et al. Dietary cis-9, trans-11-conjugated linoleic acid reduces amyloid β-protein accumulation and upregulates anti-inflammatory cytokines in an Alzheimer's disease mouse model. Sci Rep 11 (2021): 9749.
- Maia MR, Chaudhary LC, Bestwick CS, Richardson AJ, McKain N, Larson TR, et al. Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol 10 (2010): 52.
- Danics L, Schvarcz CA, Viana P, Vancsik T, Krenács T, Benyó Z, et al. Exhaustion of protective heat shock response induces significant tumor damage by apoptosis after modulated electro-hyperthermia treatment of triple-negative breast cancer isografts in mice. Cancers (Basel) 12 (2020).
- Kang H, Oh T, Bahk YY, Kim GH, Kan SY, Shin DH, et al. HSF1 Regulates mevalonate and cholesterol biosynthesis pathways. Cancers (Basel) 11 (2019).
- Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao, F, et al. MicroRNA-135b, a HSF1 target, promotes tumor invasion and metastasis by regulating RECK and EVI5 in hepatocellular carcinoma. Oncotarget 6 (2015): 2421-2433.
- Lindén SK, Sheng YH, Every AL, Miles KM, Skoog EC, Florin TH, et al. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog 5 (2009): e1000617.
- Wang YS, Zhu H, Li H, Li Y, Zhao B, Jin YH. Ginsenoside compound K inhibits nuclear factor-kappa B by targeting Annexin A2. J Ginseng Res 43 (2019): 452-459.
- Islam M, Mezencev R, McFarland B, Brink H, Campbell B, Tasadduq B, et al. Microfluidic cell sorting by stiffness to examine heterogenic responses of cancer cells to chemotherapy. Cell Death Dis 9 (2018): 239.
- Teng YN, Wang CCN, Liao WC, Lan YH, Hung CC. Caffeic Acid Attenuates multidrug resistance in cancer cells by inhibiting efflux function of human p-glycoprotein. Molecules 25 (2020).
- Chen Y, Wei L, Zhang X, Liu X, Chen Y, Zhang S, et al. 3-Bromopyruvate sensitizes human breast cancer cells to TRAIL-induced apoptosis via the phosphorylated AMPK-mediated upregulation of DR5 Corrigendum in /10.3892/or.2022.8264. Oncol Rep 40 (2018): 2435-2444.
- Bose M, Kamra M, Mullick R, Bhattacharya S, Das S, Karande AA. Identification of a flavonoid isolated from plum (Prunus domestica) as a potent inhibitor of Hepatitis C virus entry. Sci Rep 7 (2017): 3965.
- Shah NA, Khan MR, Naz K, Khan MA. Antioxidant potential, DNA protection, and HPLC-DAD analysis of neglected medicinal Jurinea dolomiaea Biomed Res Int (2014): 726241.
- Zuo G, Je KH, Guillen Quispe YN, Shin KO, Kim,HY, Kim KH, et al. Separation and identification of antioxidants and aldose reductase inhibitors in Lepechinia meyenii (Walp.) Epling. Plants (Basel) (2021).
- Liu Y, Wang D, Li Z, Li X, Jin M, Jia N, et al. Pan-cancer analysis on the role of PIK3R1 and PIK3R2 in human tumors. Sci Rep 12 (2022): 5924.
- Ai X, Xiang L, Huang Z, Zhou S, Zhang S, Zhang T, et al. Overexpression of PIK3R1 promotes hepatocellular carcinoma progression. Biol Res 51 (2018): 52.
- Gimple RC, Kidwell RL, Kim LJY, Sun T, Gromovsky AD, Wu Q, et al. Glioma stem cell-specific super-enhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov 9 (2019): 1248-1267.
- Kochupurakkal BS, Wang ZC, Hua T, Culhane AC, Rodig SJ, Rajkovic-Molek K, et al. RelA-induced interferon response negatively regulates proliferation. PLoS One 10 (2015): e0140243.
- Lomert E, Turoverova L, Kriger D, Aksenov ND, Nikotina AD, Petukhov A, et al. Co-expression of RelA/p65 and ACTN4 induces apoptosis in non-small lung carcinoma cells. Cell Cycle 17 (2018): 616-626.
- Galenkamp KM, Carriba P, Urresti J, Planells-Ferrer L, Coccia E, Lopez-Soriano J, et al. TNFα sensitizes neuroblastoma cells to FasL-, cisplatin- and etoposide-induced cell death by NF-κB-mediated expression of Fas. Mol Cancer 14 (2015): 62.
- Zheng Y, Li X, Qian X, Wang Y, Lee JH, Xia Y, et al. Secreted and O-GlcNAcylated MIF binds to the human EGF receptor and inhibits its activation. Nat Cell Biol 17 (2015): 1348-1355.
- Trippett TM, Herzog C, Whitlock JA, Wolff J, Kuttesch J, Bagatell R, et al. Phase I and pharmacokinetic study of cetuximab and irinotecan in children with refractory solid tumors: a study of the pediatric oncology experimental therapeutic investigators' consortium. J Clin Oncol 27 (2009): 5102-5108.
- Kim E, Lisby A, Ma C, Lo N, Ehmer U, Hayer KE, et al. Promotion of growth factor signaling as a critical function of β-catenin during HCC progression. Nat Commun 10 (2019): 1909.
- Yoon JH, Kim O, Eun JW, Choi SS, Ashktorab H, Smoot DT, et al. Multiple genetic mutations caused by NKX6.3 depletion contribute to gastric tumorigenesis. Sci Rep 8 (2018): 17609.
- Son SM, Park SJ, Lee H, Siddiqi F, Lee JE, Menzies FM, et al. Leucine Signals to mTORC1 via its metabolite acetyl-coenzyme a. Cell Metab 29 (2019): 192-201.
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44 (2012): 694-698.
- Hildebrandt MA, Yang H, Hung MC, Izzo JG, Huang M, Lin J, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol 27 (2009): 857-871.
- Roy S, Rizvi ZA, Clarke AJ, Macdonald F, Pandey A, Zaiss DMW, et al. EGFR-HIF1α signaling positively regulates the differentiation of IL-9-producing T helper cells. Nat Commun 12 (2021): 3182.
- Liu J, Duan Z, Guo W, Zeng L, Wu Y, Chen Y, et al. Targeting the BRD4/FOXO3a/CDK6 axis sensitizes AKT inhibition in luminal breast cancer. Nat Commun 9 (2018): 5200.
- Nath LR, Gorantla JN, Thulasidasan AK, Vijayakurup V, Shah S, Anwer S, et al. Evaluation of uttroside B, a saponin from Solanum nigrum Linn, as a promising chemotherapeutic agent against hepatocellular carcinoma. Sci Rep 6 (2016): 36318.
- Wu W, Piao H, Wu F, Han Y, An D, Wu Y, et al. Yu Jin Pulvis inhibits carbon tetrachloride-induced liver fibrosis by blocking the MAPK and PI3K/Akt signaling pathways. Am J Transl Res 11 (2019): 5998-6006.
- Yang WY, Rao PS, Luo YC, Lin HK, Huang SH, Yang JM, et al. Omics-based investigation of diet-induced obesity synergized with HBx, Src, and p53 mutation accelerating hepatocarcinogenesis in zebrafish model. Cancers (Basel) 11 (2019).
- Wei W, Chua MS, Grepper S, So SK. Blockade of Wnt-1 signaling leads to anti-tumor effects in hepatocellular carcinoma cells. Mol Cancer 8 (2009): 76.
- Yu H, Yue X, Zhao Y, Li X, Wu L, Zhang C, et al. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat Commun 5 (2014): 5218.