Role of Microbiota in Atopic Dermatitis and Bronchial Asthma - Triangular Cross-Talk among Skin, Airway and Gut
Article Information
Yukihiko Kato1*, Yasuhiro Matsumura2
1Department of Dermatology, Tokyo Medical University Hachioji Medical Center, 1163, Tate-machi, Hachioji-shi, Tokyo 193-0998, Japan
2Department of Internal Medicine, Akishima Hospital, 1260 Nakagami-cho, Akishima-shi, Tokyo 196-0022, Japan
*Corresponding author: Yukihiko Kato, Department of Dermatology, Tokyo Medical University Hachioji Medical Center, 1163, Tate-machi, Hachioji-shi, Tokyo 193-0998, Japan
Received: 18 January 2020; Accepted: 23 January 2020; Published: 29 May 2020
Citation: Yukihiko Kato, Yasuhiro Matsumura. Role of Microbiota in Atopic Dermatitis and Bronchial Asthma - Triangular Cross-Talk among Skin, Airway and Gut. Archives of Clinical and Biomedical Research 4 (2020): 169-183.
Share at FacebookAbstract
The development of allergy is partly dependent on changes in individual’s microbiota which were interacted with the environment. Microbiota can be modulated by early-life microbial exposures, diet, antibiotics. Lower microbial diversity is pivotal factor in developing diseases.
Certain types of microorganisms are involved in a disease activity. Early life exposure to non-pathogenic Proteobacteria has a protective effects in developing allergies. Later, bacterias, involving Staphylococcus aureus (S.aureus) in the skin or pathogenic Proteobacteria in the airway, affects patients with atopic dermatitis (AD) and bronchial asthma (BA) respectively. Similarly Acinetobacteria in early exposure protectively effect BA. The pathogenic role of Proteobacteria phylum might differ between bronchial and skin inflammation. The microbiota at local sites is also involved in the development and activity of diseases in remote organs via‘triangular cross talk’. Cross talk among skin, air way, and gut is not surprising, because they are the superficial organs.
Lactobacilus in the Firmicutis phylum always protectively work for allergic diseases of skin and bronchus. Therefore probiotics, which mature the gut barrier and prime the immune function, are currently being used to prevent and treat AD and BA. The accumulated data, however, have failed to substantiate fully the effects of probiotics against allergic disorders.
Keywords
Atopic dermatitis; Bronchial asthma; Microbiome; Probiotics; Proteobacteria phylum
Atopic dermatitis articles, Bronchial asthma articles, Microbiome articles, Probiotics articles, Proteobacteria phylum articles
Article Details
Introduction
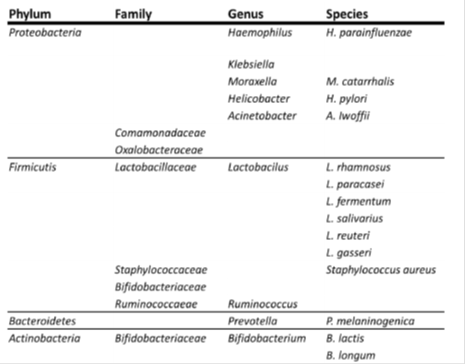
Not surprisingly, the collective genomes of the microbiota, or the microbiome, in the human body influence the prevention and development of diseases, since the genetic repertoire of the healthy microbiome is approximately 100 times that of the human genome [1]. Reviews in the past were often discussed from the point of the development of the microbiomes and diseases, based on the hygene hypothesis. In this review the relationship between microbiomes (Table 1) and allergic diseases was discussed clinically, especially the association among remote organs.
The taxonomy shows the phyla, families, genera, and species according to the International Conference on Genomics (ICG).
Environmental exposure to microorganism and allergic diseases
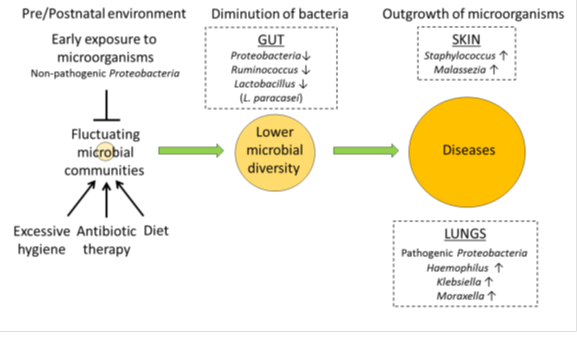
Early environmental exposure to microorganisms [2], antibiotic therapy [3] [4] [5] [6] [7], and diet [8] [9] [10], influence the composition of this microbiota. The gut microbiota is crucial to the development and maturation of the host immune system [11]. A lack of early microbial stimulation provokes an aberrant immune response to antigens leading to the development of allergies [12] (Figure 1).
Immunological changes occur at mucosal surfaces of gut due to extensive microbial colonization in early lifetime. Microbiota can be modulated by early-life microbial exposures, diet, antibiotics. Reduced diversity of gut microbial is associated with risks of allergic diseases. Certain types of microorganisms are involved in a disease activity.
Early exposure to a rural or agricultural environment influences the development of allergic symptoms later in life via microbial exposure, which elicits long-lasting effects on invariant natural killer T (iNKT) cells [13]. Acinetobacter lwoffii F78 and Lactococcus lactis G121, isolated in cowsheds of farms, were able to induce a T-helper 1 (Th1)-polarizing program in dendritic cells in mice [14] [15]. It has also been shown that Acinetobacter lwoffii F78 confers a protective effect against allergy via Toll-like receptor (TLR) signaling [16]. A recent study indicated that a lower prevalence of allergies was associated with a greater abundance and diversity of bacteria of the genus Acinetobacter at the skin and nasal mucosa in children [17].
Neonatal gut with less Bifidobacteriaceae and Lactobacillaceae, and higher Candida and Rhodotorula, is a risk of allergic diseases [18]. A particular microbiota in gut shift immune development toward Th17 responses, resulting in allergy later in childhood [19].
Skin microbiota influences AD
The skin acts not only as a physical, but also as an immunological barrier to the external environment. Human skin houses on average one million bacteria per square centimeter, which may play a role in modulating allergic disorders of the skin [20] by priming resident T lymphocyte function [21] [22] and the homeostasis between Th1 and Th2 cells, resulting in anti-inflammatory responses to environmental allergens. Although a preceding Th2-skewed immune response has not been clearly demonstrated, the absence of microbiota enhances thymic stromal lymphopoietin (TSLP) expression in mice with a defective skin barrier [23].
Microbes are thought to communicate with the cells of the surrounding skin or subcutaneous tissue. A recent study demonstrated that bacteria exist not only on the surface of the epidermis but normally also in the various layers of the dermis and subcutaneous tissue. This astonishing study suggested that bacteria could penetrate the skin barrier and interact with a wide variety of cells of the epidermis, dermis, and adipose tissue [24]. Adipocytes respond to invasive S. aureus by producing the antimicrobial peptide, cathelicidin, suggesting that subcutaneous adipocytes produce an important host defense response against skin infections [25].
Normal skin microbial flora mainly consist of the bacterial phyla, Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria [26][27]. The microbial community at disease sites of AD patients was dramatically different from that of controls. Although infants with AD were not colonized with Staphylococcus aureus (S. aureus) before the development of AD [28], colonization of AD skin by S. aureus has been correlated to an increase in disease severity [29] [30] [31] (Figure1). However, a causal association has not been established since the elimination of S. aureus does not constitute a cure for AD. On the other hand, Acinetobacter Iwoffii. induced Th1 and anti-inflammatory responses in immune and skin cells and protected against allergic sensitization and lung inflammation by way of the skin [32]. The recovery of bacterial diversity is apparently related to the achievement of remission via treatment, suggesting that the loss of microbial diversity may contribute to the pathogenesis of AD [26] [33]. Early gut microflora precede the later development of atopic sensitization [56]. Studies focusing oncorrelations between reduced early-life gut microbial diversity and elevated risks of eczema have been reported [57] [58]. A relatively diminished population of bacteria including Ruminococcus and Proteobacteria was associated with development of IgE-associated eczema [59] (Figure1). The colonization by L. paracasei decreased the risk of AD development [60].
Fungi are also part of the commensal flora in all body sites. Yeasts of the genus Malassezia in particular are associated with AD. Unlike healthy individuals, a high proportion of AD patients are sensitized to Malassezia spp. [34] (Figure1).
Airway microbiota influences bronchial asthma
The formation of the airway microbiota plays an important role in regulatory cells induction early in life [35]. During infancy an excessively "hygienic" environment decreases bacterial diversity in the airway, leading to increased susceptibility to allergic diseases [36].
The microbiota of asthmatic airways is often disturbed or altered [37] [38] and may modulate inflammatory processes in patients with severe BA and related phenotypes. A greater prevalence of Proteobacteria in the bacterial composition of the airway has been reported among patients with asthma. Haemophilus species were much more frequent in the bronchi of adult asthmatics [39], who also showed an increasing enrichment in Klebsiella spp. with increasing severity of the disease [40]. An altered upper airway microbiota characterized by lower microbial diversity and a preponderance of genus Moraxella was associated with asthma [41]. The presence of the Comamonadaceae, Sphingomonadaceae, and Oxalobacteraceae, in patients with BA correlated with bronchial hyper-responsiveness [42]. As well, the abundance of Moraxella catarrhalis or Haemophilus spp. correlated with severer pulmonary dysfunction and a higher sputum IL-8 concentration and neutrophil count [43].
Glucocorticoid (GC)-resistance is a major barrier in managing bronchial asthma [44] and requires a novel therapeutic strategy. GC responsiveness has been linked to the airway bacterial microbiome in the following manner [45]: altered airway microbiome composition stimulates airway cells, resulting in a reduced cellular response to GC. Haemophilus parainfluenzae and Prevotella melaninogenica inhibit GR-mediated mitogen-activated kinase phosphatase 1 (MKP-1) production, which dephosphorylates activated p38 mitogen-activated protein kinase via activation of transforming growth factor-β–associated kinase-1 (TAK1) and suppresses GR inhibition of NF-κB–induced IL-8 production in monocytes/macrophages [46]. Furthermore, pulmonary exposure to Escherichia coli, resulted in a protective effect against Th2-associated allergic responses [47].
Link between gut microbiota with AD and BA
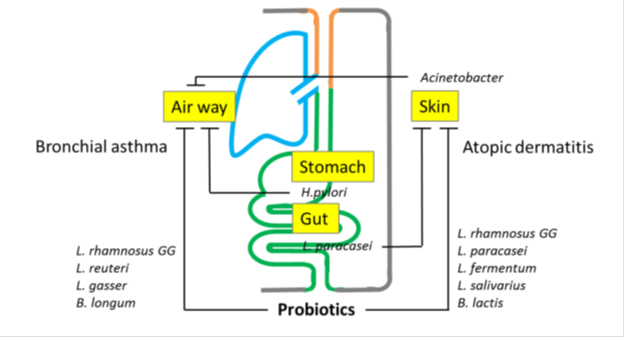
The host’ s disease is reflected by the composition of microorganisms inhabiting local sites, skin and lung. Disturbances in the microbiota result in an imbalanced immune system with consequent susceptibility to AD and BA (Figure 2). Signals from microbes can influence the cell-mediated immune system and allergies through phenotypic changes in Dendric cells (DCs), promoting regulatory T (Tregs), Th1, and natural killer (NK) cells, which inhibit Th2 inflammation [48]. Commensal-derived signals were found to influence basophil development by limiting proliferation of bone marrow-resident precursor populations, indicating that basophils are an important link between the gut microbiota and Th2 cytokine-dependent inflammation and allergic disease [49].
The host’ s disease is reflected by the composition of microorganisms inhabiting local sites. Disturbances in the microbiota result in an imbalanced immune system with consequent susceptibility to AD and BA. The microbiota at local sites is also involved in the development and activity of diseases in remote organs via ‘triangular cross talk’. Microbiota in the gut, skin and lungs can influence each other in the inception and progress of diseases.
Lactobacillus plantarum (L. plantarum) crosstalks to intestine DCs via its encypted pepide, which expands the production of regulatory IL-10 [50]. Bacteroides fragilis [51] and Clostridium strains [52] [53] can promote Treg activity to induce mucosal tolerance in the intestine. Responses to microbial components and products are key to protect indivisuals from developing allergic diseases. Bacterial polysaccharides activate CD4(+)Foxp3(-) T cells upon exposure in the gut and facilitate resistance to unnecessary inflammatory responses via the production of IL-10 [54]. Capsular polysaccharide A, produced on the surface of Bacteroides fragilis, promotes production of Th1 cytokines [55].
While direct airway microbiome manipulation influences airway immune responses, the gut microbiome has a demonstrable effect on the pathogenesis of BA similar to that seen in AD. Several studies using animal models have identified an association between alterations in the composition of gut bacterial communities and the development of BA. Germ-free mice exhibited an increase in airway eosinophils, Th2 cytokine production, IgE, and altered numbers and phenotypes of DCs after sensitization with ovalbumin (OVA). Recolonization with complex commensal flora abolished the phenotype, suggesting that the presence of commensal bacteria was critical for allergic airway inflammation [61]. Mice infected neonatally with Helicobacter pylori demonstrated greater protection against asthma [62], supporting the “disappearing microbiota” hypothesis [63], which postulates that the loss or disappearance of our ancestral indigenous microflora, rather than a general decline in arbitrary childhood infections, is associated with asthma epidemics. Helminths promote remote, protective, antiviral effects in the lung through induction of a microbiota-dependent type I IFN response [65]. Fermentable dietary fiber and short-chain fatty acids (SCFAs), its metabolites, can shape the immunological environment in the lungs, suggesting that the metabolic product of intestinal microbiota may dampen allergic responses in the lungs [66].
Fungal and bacterial microbiota during the first 100 days of life is important in the development of atopic wheeze [67]. Gut fungi are involved in promoting allergic inflammations [68]. Colonization of mice by Candida albicans following broad-spectrum antibiotic therapy promoted the development of allergic airway disease [69] [70]. Recent high dietary intake of sugar and carbohydrates and frequent use of antibiotics may be associated with Candida overgrowth in the gut. Gut fungal overgrowth promotes allergic airway inflammation.by elevated plasma PGE2 that promoted M2 macrophage polarization [71]. The microbiota at local sites is also involved in the development and activity of diseases in remote organs via triangular cross talk.’ Microbiota in the gut, skin and lungs can influence each other in the inception and progress of diseases (Figure 2). Further studies should be expected about the association between the function and microbiomes.
The effect of probiotics on allergic disorders
Bacteria-host interactions may bring about beneficial changes in immune responses. Probiotics, defined as live micro-organisms introduced into the body, promote the enrichment of regulatory dendritic cells (rDCs) and Tregs in areas of inflammation. An increase in CD4+Foxp3+ Tregs in the mesenteric lymph node (MLN) after administration of probiotics was observed in a murine model [72]. Probiotics also enhanced the secretion of interleukin (IL)-10 and Foxp3 expression in the peripheral blood of humans [73].
In mice, heat-treated Lactobacillus rhamnosus GG (L. GG) may be able to delay the onset and suppress the development of atopic dermatitis, probably through strong induction of IL-10 systemically, and in the intestinal lymphoid organs, in mice [74], and was effective in preventing early atopic disease in children with a high risk of AD development [75]. Administration of L. GG to infants with a high risk of atopy and/or their mothers appeared effective in preventing AD development [76].
A combination of L. rhamnosus and Bifidobacteria lactis improved AD in food-sensitized children [77]. Clinical improvement was reported in children with AD after they were given a mixture of L. paracasei and L. fermentum [78]. Probiotic L fermentum VRI-003 PCC is beneficial for improving the extent and severity of AD in young children with a moderate or severe form of the disease [79]. L. salivarius LS01 (DSM 22775) improved the quality of life of children affected by AD [80], and its potential usefulness in treating adult patients with AD [81] has also been reported. A meta-analysis examined randomized (placebo) controlled trials (RCTs) investigating the efficacy of probiotics in the management and prevention of AD in comparison with a placebo [82] [83] [84] [85]. Currently available evidence does not indicate that probiotic supplementation reduces the risk of allergies developing in children. However, the WAO guideline panel has suggested conditional recommendations supported by very low quality evidence [86].
Probiotics have been proposed as a therapeutic agent for BA because it is known that the generation of Treg cells is one of the mechanisms by which probiotics suppress inflammation in asthma [87] [88].
In BALB/c mice pretreated with L.GG [87] [88] [89] [90], Lactobacillus reuteri (L. reuteri) [91] [92], L. gasseri [93], and Bifidobacterium longum [94] [95], attenuated asthmatic responses induced by allergen challenges have been reported, suggesting a preventative effect. However, the results of randomized controlled trials investigating the therapeutic effectiveness of probiotics in patients with BA are not yet available.
Conclusions
The skin, as the outer surface of the body, and the lungs and the gastrointestinal tract, which comprise the inner surfaces, are the predominant sites of microbial contact. Diversity of the commensal bacteria protects against AD and BA, and alterations in the microbiota of affected tissues and intestine influence the disease state. The influence of bacteria on diseases is age-sensitive, and their role varies among species and affected sites. Modulating the gut microbiota composition is a promising strategy for treating AD and BA. Unfortunately, the findings accumulated thus far are too variable to allow hard and fast conclusions to be drawn as to the effect of probiotic use in AD and BA.
Author contributions
Yukihiko Kato wrote the AD part and Yasuhiro Matsumura did BA part mainly.
Disclosure
All authors reports no conflicts of interest in this work.
References
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124 (2006): 837-848.
- Riedler J, Braun-Fahrländer C, Eder W, Schreuer M, Waser M, Maisch S, Carr D, Schierl R, Nowak D, von Mutius E; ALEX Study Team. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet 358 (2001): 1129-1133.
- Wickens K, Pearce N, Crane J, Beasley R. Antibiotic use in early childhood and the development of asthma. Clin Exp Allergy 29 (1999): 766-771.
- Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 13 (2012): 440-447.
- Russell SL, Gold MJ, Reynolds LA,et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J Allergy Clin Immunol 135 (2015): 100-109.
- Mulder B, Pouwels KB, Schuiling-Veninga CC, et al. Antibiotic use during pregnancy and asthma in preschool children: the influence of confounding. Clin Exp Allergy 46 (2016): 1214-1226.
- Timm S, Schlünssen V, Olsen J, Ramlau-Hansen CH. Prenatal antibiotics and atopic dermatitis among 18-month old children in the Danish National Birth Cohort. Clin Exp Allergy 47 (2017): 929-936.
- Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341 (2013): 569-573.
- Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504 (2013): 446-450.
- Furusawa Y, Obata Y, Hase K. Commensal microbiota regulates T cell fate decision in the gut. Semin Immunopathol 37 (2015): 17-25.
- Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol 159 (2015):122-127.
- Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis 1 (2001): 69-75.
- Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336 (2012): 489-493.
- Debarry J, Garn H, Hanuszkiewicz A,et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol 119 (2007): 1514-1521.
- Debarry J, Hanuszkiewicz A, Stein K, Holst O, Heine H. The allergy-protective properties of Acinetobacter lwoffii F78 are imparted by its lipopolysaccharide. Allergy 65 (2010): 690-697.
- Conrad ML, Ferstl R, Teich R, et al. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med 206 (2009): 2869-2877.
- Ruokolainen L, Paalanen L, Karkman A, et al. Significant disparities in allergy prevalence and microbiota between the young people in Finnish and Russian Karelia. Clin Exp Allergy 47 (2017): 665-674.
- Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22 (2016): 1187-1191
- Petursdottir DH, Nordlander S, Qazi KR, Carvalho-Queiroz C, Ahmed Osman O, Hell E, Björkander S, Haileselassie Y, Navis M, Kokkinou E, Lio IZL, Hennemann J, Brodin B, Huseby DL, Nilsson C, Hughes D, Udekwu KI, Sverremark-Ekström E. Early-Life Human Microbiota Associated With Childhood Allergy Promotes the T Helper 17 Axis in Mice. Front Immunol 8 (2017): 1699.
- Grice EA, Segre JA. The skin Nat Rev Microbiol 9 (2011): 244-253.
- Naik S, Bouladoux N, Wilhelm C, et al. Compartmentalized control of skin immunity by resident commensals. Science 337 (2012): 1115-1119.
- Kosiewicz MM, Zirnheld AL, Alard P. Tuning of skin immunity by skin commensal bacteria. Immunotherapy 5 (2013): 23-25.
- Yockey LJ, Demehri S, Turkoz M, et al. The absence of a microbiota enhances TSLP expression in mice with defective skin barrier but does not affect the severity of their allergic inflammation. J Invest Dermatol 133 (2013): 2714-2721.
- Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun4 (2013): 1431.
- Zhang LJ, Guerrero-Juarez CF, Hata T, et al. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 347 (2015): 67-71.
- Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22 (2012): 850-859.
- Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep 15 (2015): 65.
- Kennedy EA, Connolly J, Hourihane JO, et al. Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 139 (2017): 166-172.
- Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol 90 (1974): 525-530.
- Hanifin JM, Rogge JL. Staphylococcal infections in patients with atopic dermatitis. Arch Dermatol 113 (1977): 1383-1386.
- Jun SH, Lee JH, Kim SI, et al. Staphylococcus aureus-derived membrane vesicles exacerbate skin inflammation in atopic dermatitis. Clin Exp Allergy 47 (2017): 85-96.
- Fyhrquist N, Ruokolainen L, Suomalainen A, et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J Allergy Clin Immunol 134 (2014): 1301-1309.
- Seite S, Flores GE, Henley JB, et al. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J Drugs Dermatol 13 (2014): 1365-1372.
- Johansson C, Sandström MH, Bartosik J, et al. Atopy patch test reactions to Malassezia allergens differentiate subgroups of atopic dermatitis patients. Br J Dermatol 148 (2003): 479-488.
- Gollwitzer ES, Saglani S, Trompette A, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med 20 (2014): 642-647.
- Yu W, Yuan X, Xu X, et al. Reduced airway microbiota diversity is associated with elevated allergic respiratory inflammation. Ann Allergy Asthma Immunol 115 (2015): 63-68.
- Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol 131 (2013): 346-352.e1-3.
- Pérez-Losada M, Alamri L, Crandall KA, Freishtat RJ. Nasopharyngeal Microbiome Diversity Changes over Time in Children with Asthma. PLoS One 12 (2017): e0170543.
- Hilty M, Burke C, Pedro H, et al.. Disordered microbial communities in asthmatic airways. PLoS One 5 (2010): e8578.
- Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol 136 (2015): 874-884.
- Depner M, Ege MJ, Cox MJ, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol 139 (2017): 826-834.e13.
- Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol 127 (2011): 372-381.e1-3.
- Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentiallypathogenicairway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One 9 (2014): e100645.
- Matsumura Inflammatory cellular phenotypes and molecular mechanisms of glucocorticoid resistance in patients with bronchial asthma. Antiinflamm Antiallergy Agents Med Chem 12 (2013): 189-200.
- Durack J, Lynch SV, Nariya S, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol 140 (2017): 63-75.
- Goleva E, Jackson LP, Harris JK, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med 188 (2013): 1193-1201.
- Nembrini C, Sichelstiel A, Kisielow J, Kurrer M, Kopf M, Marsland BJ. Bacterial-induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax 66 (2011): 755-763.
- Ferreira CM, Vieira AT, Vinolo MA, Oliveira FA, Curi R, Martins Fdos S. The central role of the gut microbiota in chronic inflammatory diseases. J Immunol Res 2014 (2014): 689492.
- Hill DA, Siracusa MC, Abt MC, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med 18 (2012): 538-546.
- Bernardo D, Sánchez B, Al-Hassi HO, et al. Microbiota/host crosstalk biomarkers: regulatory response of human intestinal dendritic cells exposed to Lactobacillus extracellular encrypted peptide. PLoS One 7 (2012): e36262.
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci 107 (2010): 12204-12209.
- Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331 (2011): 337-341.
- Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature500 (2013): 232-236.
- Johnson JL, Jones MB, Cobb BA. Bacterial capsular polysaccharide prevents the onset of asthma through T-cell activation. Glycobiology 25 (2015): 368-375.
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122 (2005): 107-118.
- Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol 107 (2001): 129-134.
- Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S, Coates AR, Hesselmar B, Saalman R, Molin G, Ahrné S. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol 121 (2008): 129-134..
- Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol 129 (2012):434-440.
- West CE, Rydén P, Lundin D, Engstrand L, Tulic MK, Prescott SL. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy 45 (2015): 1419-1429.
- Penders J, Thijs C, Mommers M, et al. Intestinal lactobacilli and the DC-SIGN gene for their recognition by dendritic cells play a role in the aetiology of allergic manifestations. Microbiology 156 (2010): 3298-3305.
- Herbst T, Sichelstiel A, Schär C, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med 184 (2011): 198-205.
- Arnold IC, Dehzad N, Reuter S, et al. Helicobacter pyloriinfection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest 121 (2011): 3088-3093.
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol 7 (2009): 887-894.
- Wilson MS, Taylor MD, Balic A, Finney CA, Lamb JR, Maizels RM. Suppression of allergic airway inflammation by helminth-induced regulatory T cells. J Exp Med 202 (2005): 1199-1212.
- McFarlane AJ, McSorley HJ, Davidson DJ, et al. Enteric helminth-induced type-I interferon signalling protects against pulmonary virus infection through interaction with the microbiota. J Allergy Clin Immunol 140 (2017): 1068-1078.
- Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20 (2014): 159-166.
- Arrieta MC, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, Vaca M, Boutin RCT, Morien E, Jin M, Turvey SE, Walter J, Parfrey LW, Cooper PJ, Finlay B. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 31 (2017): 649-4.
- Goldman DL, Huffnagle GB. Potential contribution of fungal infection and colonization to the development of allergy. Med Mycol 47 (2009): 445-456.
- Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72 (2004): 4996-5003.
- Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun 73 (2005): 30-38.
- Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Núñez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2.Cell Host Microbe 15 (2014): 95-102
- Kwon HK, Lee CG, So JS, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci U S A 107 (2010): 2159-2164.
- Konieczna P, Groeger D, Ziegler M et al. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: potential role for myeloid and plasmacytoid dendritic cells. Gut 61 (2012): 354-366.
- Sawada J, Morita H, Tanaka A, Salminen S, He F, Matsuda H. Ingestion of heat-treated Lactobacillus rhamnosus GG prevents development of atopic dermatitis in NC/Nga mice. Clin Exp Allergy 37(2007): 296-303.
- Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357 (2001): 1076-1079.
- Rutten NB, Gorissen DM, Eck A, et al. Long Term Development of Gut Microbiota Composition in Atopic Children: Impact of Probiotics. PLoS One 10 (2015): e0137681.
- Sistek D, Kelly R, Wickens K, Stanley T, Fitzharris P, Crane J. Is the effect of probiotics on atopic dermatitis confined to food sensitized children? Clin Exp Allergy 36 (2006): 629-633.
- Wang IJ, Wang JY. Children with atopic dermatitis show clinical improvement after Lactobacillus exposure. Clin Exp Allergy 45 (2015): 779-787.
- Weston S, Halbert A, Richmond P, Prescott SL. Effects of probiotics on atopic dermatitis: a randomised controlled trial. Arch Dis Child 90 (2005): 892-897.
- Niccoli AA, Artesi AL, Candio F, et al..Preliminary results on clinical effects of probiotic Lactobacillus salivarius LS01 in children affected by atopic dermatitis. J Clin Gastroenterol 48 (2014): S34-36.
- Drago L, De Vecchi E, Toscano M, Vassena C, Altomare G, Pigatto P. Treatment of atopic dermatitis eczema with a high concentration of Lactobacillus salivarius LS01 associated with an innovative gelling complex: a pilot study on adults. J Clin Gastroenterol 48 (2014): S47-51.
- Betsi GI, Papadavid E, Falagas ME. Probiotics for the treatment or prevention of atopic dermatitis: a review of the evidence from randomized controlled trials. Am J Clin Dermatol 9 (2008): 93-103.
- Pelucchi C, Chatenoud L, Turati F, et al. Probiotics supplementation during pregnancy or infancy for the prevention of atopic dermatitis: a meta-analysis. Epidemiology 23 (2012): 402-414.
- Panduru M, Panduru NM, Sălăvăstru CM, Tiplica GS. Probiotics and primary prevention of atopic dermatitis: a meta-analysis of randomized controlled studies. J Eur Acad Dermatol Venereol 29 (2015): 232-242.
- Kim SO, Ah YM, Yu YM, Choi KH, Shin WG, Lee JY. Effects of probiotics for the treatment of atopic dermatitis: a meta-analysis of randomized controlled trials. Ann Allergy Asthma Immunol 113 (2014): 217-226.
- Fiocchi A, Pawankar R, Cuello-Garcia C, et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J 8 (2015): 4.
- Feleszko W, Jaworska J, Rha RD, et al. Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin Exp Allergy 37 (2007): 498-505.
- Jang SO, Kim HJ, Kim YJ,et al. Asthma Prevention by Lactobacillus Rhamnosus in a Mouse Model is Associated With CD4(+)CD25(+)Foxp3(+) T Cells. Allergy Asthma Immunol Res 4 (2012): 150-156.
- Blümer N, Sel S, Virna S, et al. Perinatal maternal application of Lactobacillus rhamnosus GG suppresses allergic airway inflammation in mouse offspring. Clin Exp Allergy 37 (2007): 348-357.
- Yu J, Jang SO, Kim BJ, et al. The Effects of Lactobacillus rhamnosus on the Prevention of Asthma in a Murine Model. Allergy Asthma Immunol Res 2 (2010): 199-205.
- Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med 175 (2007): 561-569.
- Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med 179 (2009): 186-193.
- Jan RL, Yeh KC, Hsieh MH, et al. Lactobacillus gasseri suppresses Th17 pro-inflammatory response and attenuates allergen-induced airway inflammation in a mouse model of allergic Br J Nutr 108 (2012): 130-139.
- Lyons A, O'Mahony D, O'Brien F, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy 40 (2010): 811-819.
- MacSharry J, O'Mahony C, Shalaby KH, et al. Immunomodulatory effects of feeding with Bifidobacterium longum on allergen-induced lung inflammation in the mouse. Pulm Pharmacol Ther 25 (2012): 325-334.