Re-Profiling of US-FDA Approved Drugs for COVID-19 by In-silico Studies
Article Information
Akash Parab1, Dr Gaganjyot Kaur2 *, Abhishek Sharma2 *, Gursewak Bhuller2 *, Sharvari Chitnis2 *, Ankit Chindaliya2 *, Mayur Wetal2 *
1Guru Nanak Institute of Research and Development, India
2Guru Nanak Khalsa College of Arts, Science and Commerce, Nathalal Parekh Marg, Matunga East, Mumbai – 400019 Maharashtra, India
*Corresponding Authors: Dr. Gaganjyot Kaur, Head GNIRD, Guru Nanak Khalsa College of Arts, Science and Commerce, Nathalal Parekh Marg, Matunga East, Mumbai – 400019 Maharashtra, India
Received: 15 June 2021; Accepted: 24 June 2021; Published: 29 June 2021
Citation: Akash Parab, Dr Gaganjyot Kaur, Abhishek Sharma, Gursewak Bhuller, Sharvari Chitnis, Ankit Chindaliya, Mayur Weta. Re-Profiling of US-FDA Approved Drugs for COVID-19 by In-silico Studies. Journal of Bioinformatics and Systems Biology 4 (2021): 50-73.
Share at FacebookAbstract
Covid-19 is an infectious disease that has rattled the world. Although several new vaccines are being administered to prevent infections, there is a large unmet need for antiviral drugs to ameliorate disease progression. In this study, structures of endogenous proteins were modelled using the genome browser and screened for molecular docking of drugs with spike protein of SARS COV-2 binding to ACE2 receptor using bioinformatics approaches such as structure-assisted drug design (SBDD), virtual drug screening (VS), and high-throughput screening (HTS). We have screened 150 drugs that were shortlisted from extensive literature data, 110 of these have been approved by the US-FDA for various diseases. We have developed a molecule assessment process from which we have identified fifteen drugs that have the potential for repurposing towards blocking SARS-COV2 proteins with endogenous proteins. Using these approaches, we have identified asthma drugs such as Cromolyn sodium, Ciclesonide and Zafirlukast.
Keywords
COVID-19; Drugs; In-silico analysis; Repurposing of drugs; USFDA; Structure assisted drug design
COVID-19 articles; Drugs articles; In-silico analysis articles; Repurposing of drugs articles; USFDA articles; Structure assisted drug design articles
COVID-19 articles COVID-19 Research articles COVID-19 review articles COVID-19 PubMed articles COVID-19 PubMed Central articles COVID-19 2023 articles COVID-19 2024 articles COVID-19 Scopus articles COVID-19 impact factor journals COVID-19 Scopus journals COVID-19 PubMed journals COVID-19 medical journals COVID-19 free journals COVID-19 best journals COVID-19 top journals COVID-19 free medical journals COVID-19 famous journals COVID-19 Google Scholar indexed journals Drugs articles Drugs Research articles Drugs review articles Drugs PubMed articles Drugs PubMed Central articles Drugs 2023 articles Drugs 2024 articles Drugs Scopus articles Drugs impact factor journals Drugs Scopus journals Drugs PubMed journals Drugs medical journals Drugs free journals Drugs best journals Drugs top journals Drugs free medical journals Drugs famous journals Drugs Google Scholar indexed journals In-silico analysis articles In-silico analysis Research articles In-silico analysis review articles In-silico analysis PubMed articles In-silico analysis PubMed Central articles In-silico analysis 2023 articles In-silico analysis 2024 articles In-silico analysis Scopus articles In-silico analysis impact factor journals In-silico analysis Scopus journals In-silico analysis PubMed journals In-silico analysis medical journals In-silico analysis free journals In-silico analysis best journals In-silico analysis top journals In-silico analysis free medical journals In-silico analysis famous journals In-silico analysis Google Scholar indexed journals Repurposing of drugs articles Repurposing of drugs Research articles Repurposing of drugs review articles Repurposing of drugs PubMed articles Repurposing of drugs PubMed Central articles Repurposing of drugs 2023 articles Repurposing of drugs 2024 articles Repurposing of drugs Scopus articles Repurposing of drugs impact factor journals Repurposing of drugs Scopus journals Repurposing of drugs PubMed journals Repurposing of drugs medical journals Repurposing of drugs free journals Repurposing of drugs best journals Repurposing of drugs top journals Repurposing of drugs free medical journals Repurposing of drugs famous journals Repurposing of drugs Google Scholar indexed journals USFDA articles USFDA Research articles USFDA review articles USFDA PubMed articles USFDA PubMed Central articles USFDA 2023 articles USFDA 2024 articles USFDA Scopus articles USFDA impact factor journals USFDA Scopus journals USFDA PubMed journals USFDA medical journals USFDA free journals USFDA best journals USFDA top journals USFDA free medical journals USFDA famous journals USFDA Google Scholar indexed journals Structure assisted drug design articles Structure assisted drug design Research articles Structure assisted drug design review articles Structure assisted drug design PubMed articles Structure assisted drug design PubMed Central articles Structure assisted drug design 2023 articles Structure assisted drug design 2024 articles Structure assisted drug design Scopus articles Structure assisted drug design impact factor journals Structure assisted drug design Scopus journals Structure assisted drug design PubMed journals Structure assisted drug design medical journals Structure assisted drug design free journals Structure assisted drug design best journals Structure assisted drug design top journals Structure assisted drug design free medical journals Structure assisted drug design famous journals Structure assisted drug design Google Scholar indexed journals SARS-COCV-2 articles SARS-COCV-2 Research articles SARS-COCV-2 review articles SARS-COCV-2 PubMed articles SARS-COCV-2 PubMed Central articles SARS-COCV-2 2023 articles SARS-COCV-2 2024 articles SARS-COCV-2 Scopus articles SARS-COCV-2 impact factor journals SARS-COCV-2 Scopus journals SARS-COCV-2 PubMed journals SARS-COCV-2 medical journals SARS-COCV-2 free journals SARS-COCV-2 best journals SARS-COCV-2 top journals SARS-COCV-2 free medical journals SARS-COCV-2 famous journals SARS-COCV-2 Google Scholar indexed journals US-FDA articles US-FDA Research articles US-FDA review articles US-FDA PubMed articles US-FDA PubMed Central articles US-FDA 2023 articles US-FDA 2024 articles US-FDA Scopus articles US-FDA impact factor journals US-FDA Scopus journals US-FDA PubMed journals US-FDA medical journals US-FDA free journals US-FDA best journals US-FDA top journals US-FDA free medical journals US-FDA famous journals US-FDA Google Scholar indexed journals proteins articles proteins Research articles proteins review articles proteins PubMed articles proteins PubMed Central articles proteins 2023 articles proteins 2024 articles proteins Scopus articles proteins impact factor journals proteins Scopus journals proteins PubMed journals proteins medical journals proteins free journals proteins best journals proteins top journals proteins free medical journals proteins famous journals proteins Google Scholar indexed journals
Article Details
1. Introduction
Coronavirus [1] disease-2019 (Covid-19) is a lethal zoonotic, contagious, global emergency pandemic. The incidences of COVID-19 infections are rapidly increasing, due to the transmission from symptomatic/asymptomatic carriers [2]. Seven Coronaviruses (229E, NL63, OC43, HKU1, SARS, MERS and, COVID-19) have been reported, where MERS and SARS-CoV have been associated with high mortality rates. In India, the mortality rate during a sudden increase in cases due to the current mutant is 1.14 %, which is lower than many other countries. The mechanism of this reduced mortality is yet to be elucidated [3-5]. Coronaviruses are a large group of viruses that causes severe respiratory infections that affect both animals and humans. It has a round or elliptic and pleomorphic form having a diameter of approximately 60–140 nm. SARS-CoV2 has a capsid of 27–32 kb, which contained a single-stranded RNA genome with positive polarity enclosed in an envelope. It contains four types of spikes; many long glycoprotein S-spikes (20 nm) in all the coronaviruses, small hemagglutinin-esterase (HE) spikes in some viruses, few transmembrane glycoproteins and envelope protein spikes [6-9]. The incidences of COVID-19 infections are rapidly increasing, due to the transmission from symptomatic/asymptomatic carriers, high mobility rate. After it enters into the host cell, the virion binds to the receptors on the target cells in the human respiratory tract. The genomic analysis, suggests that COVID-19 will utilize similar receptors that were used by SARS-CoV. The COVID-19 will be attached to the angiotensin-converting enzyme 2 (ACE-2) receptor, which is expressed in the lungs, heart, kidney, small intestine and other tissues [10-15]. The detailed life cycle of SARS-COCV-2 is shown in Figure 1.
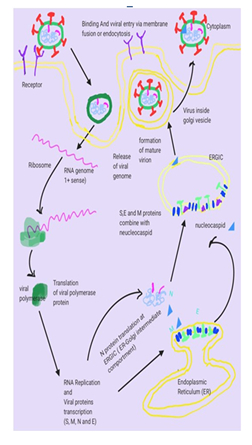
Figure1: Life cycle of SARS Coronavirus begins with Spike (S) glycoprotein binds to the cellular receptor ACE2 (Angiotensin-converting enzyme 2) which facilitates viral envelope fusion with the cell membrane through the endosomal pathway. Then SARS Coronavirus releases its genomic RNA into the host cell which is translated into viral replicase polyproteins which are cleaved into small products by viral proteinases. The polymerase produces a series of sub-genomic mRNAs and finally translated into relevant viral proteins (S, M, E and N). Viral proteins and genome RNA are subsequently assembled into virions in the ERGIC (ER–Golgi intermediate compartment) and then transported via vesicles and released out of the cell.
Coronavirus [16-20] is contagious to all age group which makes a big concern for people with comorbidities and young ones highly susceptible to infection due to lack of immune response towards n-COV-19. Most of the hospitals with the confirmed COVID-19 cases have insufficient medical facilities and where the major population is below the poverty line hence the treatment with drugs available becomes expensive. Therapeutic strategies to deal with the infection are only supportive, and prevention aimed at reducing transmission in the community is our best weapon [21-24]. The major clinically observation of covid 19 seen are on lungs which causes the c-reactive proteins level to increase, also D-dimer i.e., clots in the blood vessel, increase in inflammation (Table 1) Till now No single specific antiviral therapy for COVID-19 and the main treatments are supportive. The current study is related to find a drug that is US-FDA approved and factoring the cost of the drug for the initial line of treatment. For which drug repurposing process is used as it identifies new uses for approved or investigational drugs and it is considered as a very effective strategy for drug discovery as it involves less time and cost to find a therapeutic agent in comparison to the de novo drug discovery process [25-29].
Table 1: Currently line of treatment used for treating covid19 with its effect on patients
|
Drug |
Line of treatment with effects |
|
Hydroxychloroquine HCQs |
Mild to moderate cases, as prophylactic, but according to the European medicines Agency, clinical trial data of uses of HCQs is very limited and can lead to heart diseases |
|
Remdesivir |
Used for treatment with clinically severe illness with covid19, it blocks viral RNA polymerases activity thus reducing the viral load and alleviating pathological damage to lung tissue. It increases liver enzyme levels which possibly damage the liver, nausea, vomiting. |
|
Tocilizumab is an IgG1 monoclonal antibody |
Against the IL-6 receptor which is used in the treatment of rheumatoid arthritis |
|
Sarilumab |
Anti-IL-6 receptor antibody |
|
Anakinra |
Recombinant IL-1 receptor antagonist used to treat auto-inflammatory disorders. Analysis showed that in patients with moderate-to-severe ARDS, and hyper inflammation (C-reactive protein ≥100 mg/L, ferritin ≥ 900 ng/mL, or both), the use of anakinra induced clinical improvement in 72% of patients. |
|
Acalabrutinib |
Managing inflammation proved that the treatment improved oxygenation in many patients, ameliorating measures of inflammation such as C-reactive protein and IL-6. In some ICU patient’s thrombophilia or thrombosis |
2. Methods
There is much attention to recognize drugs that can potentially re-profile by either managing or curing covid19, for this purpose selection of pharmacological interventions was done by considering the following criteria:
- Drugs prescribed for common flu, influenza, HIV, which is mostly retroviral drugs, protease inhibitors and anti-inflammatory.
- Anti-asthma drugs as it help in improving respiration of the patient which is the most common symptom of covid-19 infections where lack of breathing occurs due to congestion of lungs and pneumonia.
- Also, these drugs should be either USFDA approved or under clinical trials were evaluated.
- To avoid the lengthy procedure of clinical trials, where 40 drugs currently being prescribed for covid-19 infections were found to lack the evidence of periodic safety update reports (PSDR), which thereby increased the chances of adverse drug reactions to critically ill patients, hence decided to include USFDA approved drugs which had been prescribed for 40 years. This narrowed down the list up to 110 drugs being confirmed for repurposing of drugs.
- Drugs with higher binding affinity were shortlisted for visualization.
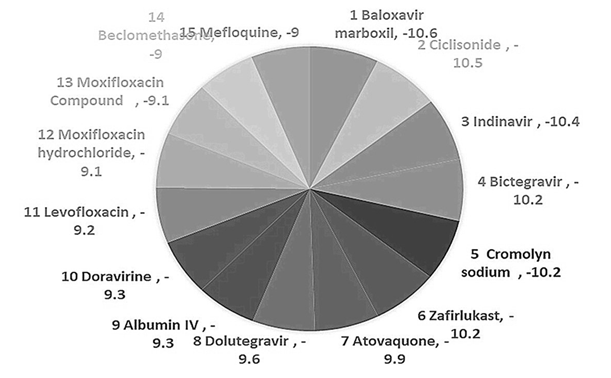
Figure 2: Pie chart of Binding affinity in Kcal/mol.
These 110 drugs (For a list of 150 drugs and 110 drugs you can refer to our special annexure 1) were further subjected to docking analysis using Auto Dock Vina 1.5.6, PyMol, and Ligplot+ v 1.4.5. Docking. Structural data obtained from protein data bank such as Pubchem were converted into PDB format using PyMol. The file format used was pdbqt using Auto Dock tools 1.5.6. Parameters of the software were tuned to default mode. The in-silico modelling of the candidate approved drugs was carried out using Autodock vina and autodock tools. The visualization of encountered ligand conformations was performed using Ligplot+, PyMoL, Protein-Ligand Interaction Profiler (PLIP).
2.1 Ligand preparation
110 USFDA approved drugs ligands were subjected to virtual screening to evaluate their potency against SARS COV-19. The 3D structures of protein counterpart’s i.e Ligands were obtained using the NCBI Pubchem database in SDS format. The ligands in SDS format were converted into PDB format using PyMol Software. These ligand conformations were prepared for docking using AutoDock tools. The prepared ligands were verified and energy minimization was carried out to obtain final docking ready ligands in pdbqt format.
2.2 Protein preparation
X-Ray Crystal Structure of the SARS Coronavirus Main Protease (pdb id -1Q2W) was acquired using Protein Data Bank. The structural improvement of the main protease was carried out by using the AutoDock tool. The water molecules present in the structure were removed and polar hydrogen was added. This was followed by merging the Non-polar hydrogen. Kollman charges were introduced which concluded the protein improvement process. The X-Ray Crystal Structure was also checked for missing residue and the entire structure was rectify by adding certain residues. Finally, the improved protein structure of the main protease was subjected to generate a Grid box using the Grid panel in the Auto-Dock tool.
2.3 Molecular docking
Molecular docking is a structure-based drug design approach to identify the essential amino acid interactions between the selected protein and generated ligands with low energy conformation with the help of AutoDock vina randomized docking protocol was implemented covering the entire stretch of X-Ray Crystal protein Structure to obtain binding affinity score in kcal/mol. The best protein-ligand conformations were carried forward for visualization.
2.4 Final visualization
PyMol, Protein-Ligand Interaction Profiler (PLIP), Ligplot+ and Protein Plus Server. Based on binding affinity to a receptor drugs will be shortlisted.
3. Results and Discussion
Intending to repurpose the drugs, we focused on in-silico repurposing of global pharmacological interventions. As the targeted virus is SARS-COV-2, which has a thick glycan coating on its surface making it difficult to breach; random docking was evaluated by its binding affinity. PDB format was used for structural data obtained from PUBCHEM. We conducted random docking from tools such as Auto Dock Vina 1.050 and Autodoc 4.2 tool. The result obtained shows the significance binding of ligands with the target protein, where binding affinity is the strength of the binding interaction between a single biomolecule i.e., protein to its ligand and the binding energy is more stable when the complex with lower energy. On this basis, drugs with ligands giving a binding affinity score of above 09 kcal/mol were selected for shortlisted and this brought us down from 110 drugs to 15 drugs with better binding affinity. We shortlisted 06 drugs with a binding affinity score of above -10 kcal/mol for visualization using Ligplot+. After visualizing the protein in PyMol it was found that SARS-COV-2 main proteases consist of A and B chains as shown in Figure 5 where both the chains are highly involved and show the interaction with drugs ligands. Please find the list of the 15 drugs that were subjected for visualization in Table 2.
Table 2: List of 15 drugs with highest binding affinity
|
Sr.No |
Drug Name |
Dosage Form |
Inference |
|
1 |
Baloxavir marboxil Mode of mechanism: CAP endonuclease inhibitor and blocks the transcription of mRNA and inactivate the influenza virus. Mode of Action: Antiviral drug used for the treatment of influenza A and influenza B infections. Generic Name: Brand Name: Xofluza Manufacturer:- Triveni Interchem Private Limited Accession No: DB13997 USFDA Approved Cost : 150USD/Strip |
20mg-40mg |
Observed Affinity -10.6 kcal/mol |
|
2 |
Ciclesonide Mode of mechanism: Inhibit leukocyte infiltration at the site of inflammation by binding to Glucocorticoid receptor. Mode of Action: (Glucocorticoid used to treat obstructive airway diseases. Generic Name: Apo-ciclesonide Brand Name: Alvesco Manufacturer:- CIPLA USFDA Approved Accession No: DB01410 Cost: - 162.24 USD/bottle |
80-160mcg/inhalation |
Observed Affinity -10.5 kcal/mol |
|
3 |
Indinavir Mode of mechanism: Inhibits the HIV viral protease enzyme which prevents cleavage of the gag-pol polyprotein which results in noninfectious, immature viral particles.
Mode of Action: A potent and specific HIV protease inhibitor used for the treatment of HIV infection Accession No: DB00224 Generic Name: Brand Name: Crixivan Manufacturer:- CIPLA USFDA Approved Accession No: DB00224 Cost: - 570.02USD/bottle |
200-400mg |
Observed Affinity -10.4 kcal/mol |
|
4 |
Bictegravir Mode of mechanism: It inhibits the integrase enzyme which prevents the integration of HIV-1 into host DNA, thus blocking the conversion of the HIV-1 provirus and progression of the virus.
Mode of Action: Bictegravir is an HIV-1 integrase strand transfer inhibitor (INSTI) which inhibits HIV-1 virus replication into the human genome. Generic Name: Brand Name: Biktarvy Manufacturer:- Taffic USFDA Approved Accession No: DB11799 Cost: - 14,315USD/25mg |
50-200mg |
Observed Affinity -10.2 kcal/mol |
|
5 |
Cromolyn sodium Mode of mechanism: It inhibits degranulation of mast cells preventing the release of histamine and slow-reacting substance SRS-A. Mode of Action: A synthetic compound that inhibits antigen-induced bronchospasms and is used to treat asthma and allergic rhinitis. Generic Name: Crolom Brand Name: Gastrocrom Manufacturer:- Cromal USFDA Approved Accession No: DB01003 Cost:- 3.92USD/ampule |
100mg/5ml |
Observed Affinity -10.2 kcal/mol |
|
6 |
Zafirlukast Mode of mechanism: Binds to Cysteinyl leukotriene receptor 1 alters the cellular activity associated with the inflammatory process. Mode of Action: Zafirlukast is an oral leukotriene receptor antagonist (LTRA) for the maintenance treatment of asthma. Accession No: DB00549 Generic Name: Zafirlukast Brand Name: Accolate Manufacturer:- Dr. Reddy's Laboratories Ltd. USFDA Approved Accession No: DB00549 Cost: - 1.92USD/Tablet |
10mg-20mg |
Observed Affinity -10.2 kcal/mol |
|
7 |
Atovaquone Mode of mechanism: Binds to cytochrome bc1 complex and mitochondrial electron transport chain resulting in inhibition of nucleic acid and ATP synthesis Mode of Action: Atovaquone is a hydroxynaphthoquinone Atovaquone is a hydroxynaphthoquinone. Generic Name: Atovaquone Brand Name: Mepron Manufacturer:- Sun Pharma USFDA Approved Accession No: DB01117 Cost:- 7.7USD/ Tablet |
250mg |
Observed Affinity -9.9 kcal/mol |
|
8 |
Dolutegravir Mode of mechanism: It inhibits HIV integrase by binding to the active site and blocking the strand transfer step of retroviral DNA integration in the host cell. Mode of Action: Dolutegravir is a HIV-1 intergrase inhibitor that blocks the strand transfer step of the integration of the viral genome into the host cell. Accession No: DB08930 Generic Name: Brand Name: Tivicay Manufacturer:- Hetero USFDA Approved Accession No: DB08930 Cost:-1,989USD/Strip |
10-50mg |
Observed Affinity -9.6 kcal/mol |
|
9 |
Darunavir + Albumin I.v Mode of mechanism: Prevents HIV replication through binding to the enzyme, stopping the dimerization and the catalytic activity of HIV-1 protease. Mode of Action: Darunavir is a protease inhibitor used with other HIV protease inhibitor drugs as well as ritonavir for the effective management of HIV-1 infection. Generic Name: Apo-darunavir Brand Name: Prezista Manufacturer:- Hetero Healthcare USFDA Approved Accession No: DB01264 Cost:-25.75USD/Tablet |
75 mg |
Observed Affinity -9.3 kcal/mol |
|
10 |
Doravirine Mode of mechanism: Doravirine inhibits HIV-1 replication by non-competitively inhibiting HIV-1 reverse transcriptase. Mode of Action: Doravirine is an HIV-1 non-nucleoside reverse transcriptase inhibitor (NNRTI) intended to be administered in combination with other antiretroviral medicines. Generic Name: Brand Name: Pifeltro Manufacturuer:- Merck Sharp & Dohme Corp. USFDA Approved Accession No: DB12301 Cost:- 1,522USD/30tablets |
100mg |
Observed Affinity -9.3 kcal/mol |
|
11 |
Levofloxacin (mode of mechanism) Inhibits DNA gyrase and topoisomerase IV which introduces negative and positive supercoils into DNA during replication thus result is a blockade of DNA replication. (mode of Action) Levofloxacin is a fluoroquinolone antibiotic is a member of the third generation fluoroquinolones which shows activity against gram-positive bacteria commonly implicated in respiratory infections. Generic Name: Apo-levofloxacin Brand Name: Act Levofloxacin Manufacturer:- Arrier Biotech USFDA Approved Accession No: DB01137 Cost:-28.06USD/Tablet. |
500mg-750mg |
Observed Affinity -9.2 kcal/mol |
|
12 |
Moxifloxacin hydrochloride Mode of mechanism: It inhibits the activity of DNA TOPOISOMERASE II antibiotic used to treat a number of bacterial infections. Mode of Action: Moxifloxacin hydrochloride is a derivative of Moxifloxacin which act as antibiotic. Generic Name: Ag-moxifloxacin Brand Name: Avelox Manufacturer:- Biomedica Remedies USFDA Approved Accession No: DB00218 Cost:- 16.68USD/Tablet |
400mg |
Observed Affinity -9.1 kcal/mol |
|
13 |
Moxifloxacin Mode of mechanism: Inhibits enzyme called DNA gyrase, which is essential for bacterial survival. (mode of Action) Moxifloxacin is a synthetic fluoroquinolone antibiotic agent. Generic Name: Ag-moxifloxacin Brand Name: Avelox Manufacturer:- Biomedica Remedies USFDA Approved Accession No: DB00218 Cost:- 16.68USD/Tablet |
5mg/ml |
Observed Affinity -9.1 kcal/mol |
|
14 |
Beclomethasone Mode of mechanism: It binds to glucocorticoid response elements (GRE) in nucleus on glucocorticoid-responsive genes, leading to changes in transcription. Mode of Action: It is a prodrug of an active metabolite beclomethasone 17-monopropionate (17-BMP) which acts on the glucocorticoid receptor to mediates its therapeutic action. Generic Name: Alti-beclomethasone Dipropionate Inhaler Brand Name: Beclodisk Manufacturer:- Connote Healthcare USFDA Approved Accession No: DB00394 Cost:- 149.32USD/Inhaler |
40-80mcg/acutation |
Observed Affinity -9.0 kcal/mol |
|
15 |
Mefloquine Mode of mechanism: It may act by forming toxic complexes with free heme that damage membranes and interact with other Viral components. Mode of Action: A phospholipid-interacting antimalarial drug which is very effective against plasmodium falciparum with very few side effects. Generic Name: Mefloquine Hydrochloride Brand Name: Lariam Manufacturer:- Zydus Cadila Healthcare Ltd. USFDA Approved Accession No: DB00358 Cost:- 12.41USD/Tablet |
250mg |
Observed Affinity -9.0 kcal/mol |
Based on the score obtained, we can infer that these drugs, such as Baloxavir marboxil, Ciclisonide, Indinavir, Bictegravir, Cromolyn sodium, Zafirlukast, were chosen for further processing based on higher binding affinity. The following section discusses docking analysis of the drugs.
- Baloxavir Marboxil being an antiviral drug used for the treatment of influenza A and influenza B infections, having its Mode of administration as Oral. Mode of mechanism, CAP endonuclease inhibitor and bloxcks the transcription of mRNA and inactivates the influenza virus. Its dose range is from 20 mg to 40 mg. Coadministration with dairy products, calcium-fortified beverages, polyvalent cation-containing laxatives or antacids, or oral supplements (e.g., calcium, iron, magnesium, selenium, zinc) should be avoided, with possible Adverse Effects1-10% such as Diarrhea (3%), Bronchitis (2%), Nausea (1%), Nasopharyngitis (1%), Headache (1%). Baloxavir Marboxil docked with SARS-COV-2 main proteases showed a significant binding with an affinity score of -10.6 kcal/mol. The interaction of Baloxavir Marboxil with the proteases showed a high-affinity interaction in chain A (Figure 4). This is further evidenced by hydrogen bonding between the oxygen of the carbonyl group of Baloxavir Marboxil with Pro 108. Some other Van der Waals interaction Baloxavir Marboxil with Pro 132, His 246, Leu 202 and Ile 200 has been observed. Baloxavir Marboxil when docked in COVID-19 proteases yielded 12 hydrophobic interactions at Arg 4, Lys 5, Phe 3, Glu 288, Ser 284 Thr 285, Ile 286 and Phe 291 of chain B while chain A showed interactions at Lys 5, Phe 3, Glu 288 and Ile 286. Ligplot visualization showed that chain B of proteases is more involved in binding with Baloxavir Marboxil at the pocket binding region.
- Bictegravir being an Antiviral drug B is an HIV-1 integrase strand transfer inhibitor (INSTI) that inhibits HIV-1 virus replication into the human genome. It inhibits the integrase enzyme which prevents the integration of HIV-1 into host DNA, thus blocking the conversion of the HIV-1 provirus and progression of the virus. Its dose range is from 20mg to 500mg. Severe acute exacerbations of hepatitis B reported in patients coinfected with HIV-1 and HBV and have discontinued products containing emtricitabine (FTC) and/or tenofovir disoproxil fumarate (TDF). Closely monitor hepatic function with both clinical and laboratory follow-up for at least several months in patients coinfected with HIV-1 and HBV who discontinued the therapy. If appropriate, anti-hepatitis B therapy may be warranted, with possible adverse effects 1-10%, Diarrhea (3-6%), Nausea (3-5%), Headache (4-5%), Creatine kinase ≥10x ULN (4%), Abnormal dreams (<3%), Fasting LDL-C >190 mg/dL (2-3%), Fatigue (2-3%), Dizziness (2%), Insomnia (2%), Neutrophil <750 mm³ (2%), Amylase >2x ULN (2%), ALT >5x ULN (1-2%), AST >5x ULN (1-2%). Bictegravir showed a high-affinity interaction with chain A and B of protein with an affinity score of -10.2 kcal/mol. The docking of Bictegravir with SARS-COV-2 main proteases revealed many types of interactions. A hydrogen bond was encountered between Bictegravir and amino acid residue of protein (Figure 5). The hydrogen bond interactions obtained are in between the A chain Lys 5 and B chain Gln 127 and Lys 5. This is followed by the alkyl interaction of benzene rings with Lys 137 of A chain. It also showed the formation of a halide bond at Val 125 of A chain and a simple carbon-hydrogen bond at Try 126. When Bictegravir was docked against COVID-19 proteases many facts were revealed. It was found that Bictegravir formed two significant hydrogen bonds with Lys 5 of chain A and Lys 5 of B chain with a bond length of 3.12 and 3.16 respectively. Hydrophobic interactions were observed at Lys 137, Gln 127 of A chain and Tyr 126, Gln 127, Phe 3, Gly 283, Gly 2, Arg 4, Gln 288 of B chain respectively.
- Ciclesonide being an Anti-Asthmatic, Anti-Inflammatory in its class of drugs, is a Glucocorticoid used to treat obstructive airway diseases is administered as an Oral Inhalation while inhibiting leukocyte infiltration at the site of inflammation by binding to Glucocorticoid receptor. Its dose range is from 80mcg to 160mcg. Prolonged use of corticosteroids leads to serious infections. Should not be used for acute asthma. High doses may cause suppression of hypothalamic-pituitary-adrenal axis, which can result in adrenal crisis. Long-term use may result in reduced BMD; glaucoma or cataracts; increased IOP Development of Kaposi's sarcoma-associated with prolonged use of corticosteroids Psychiatric disturbances reported with corticosteroid use. When Ciclesonide docked is in SARS-COV-2 main proteases shows a significant interaction in the central pocket near chain A and B with the binding affinity of -10.5 kcal/mol (Figure 6). The major interaction between Ciclesonide and the proteases is up-hold by hydrogen bonds between the oxygen group of ligands and Chain A Lys 5 and also with Arg 4 of chain B. An unfavourable acceptor interaction has also been observed at Glu 290. It also yielded a hydrogen bond connecting B chain Lys 5 with the oxygen of ligand (Ciclesonide) molecule. Some of the alkyl interactions of the aromatic ring and Ile 286 and Lys 137 have been encountered.
- Cromolyn sodium belonging to the class of Anti-inflammatory, Anti-asthmatic is administered as oral inhalation, is a synthetic compound that inhibits antigen-induced bronchospasms and is used to treat asthma and allergic rhinitis. It inhibits degranulation of mast cells preventing the release of histamine and slow-reacting substance SRS-A with a dose administered as 10mg/ml. Some reports of bronchospasm or cough after administration, not useful in acute situations, symptoms may reoccur while withdrawing or tapering the dose. Cromolyn sodium was docked with SARS-COV-2 and the results showed that the main proteases bound with Cromolyn sodium in the pocket between chain A and B with the affinity of -10.2 (Figure 7). It shows significant binding with five hydrogen bonds between oxygen and Asp 289, Arg 4 of B chain, Lys 5 and Ala 7 and Lys 5 of B chain. It also yielded a simple carbon-hydrogen bond between ligand and Phe 3 of the B chain. A pi- anion bond was also observed between the benzene ring and Glu 288 of the A chain. Two pi-alkyl and pi-sigma bonds were observed in Lys 5 and Arg 4 formation. Cromolyn sodium turns out to give 9 significant conventional hydrogen interactions between oxygen and nitrogen and oxygen present in neighbouring amino acid residues. Lys of chain B formed a hydrogen bond with 2.69 bond length and Ala 7 of chain A formed a 3.26 bond length hydrogen bond. Val 125 of A chain formed a single hydrogen bond with a bond length of 2.93. Arg of B chain formed 2 conventional hydrogen interactions with bond lengths of 3.24 and 3.01 respectively. Glu 288 of chain A developed a single hydrogen bond with a 2.73 bond length. Asp 289 of A chain formed 2 significant bonds with 2.72 and 3.15 bond lengths respectively. On the other hand, Lys 5 of chain A formed a single bond with a bond length of 3.18. A huge number of hydrophobic interactions were observed between Indinavir and proteases at Gln 127, Tyr 126, Arg 4, Ala 7, Glu 290 and Ile 286 of A chain and Gly 283, Phe3 and Met 6 of B chain respectively. Cromolyn sodium revealed a significant number of conventional hydrogen bond interactions when visualized on Ligplot.
- Indinavir being an Anti-Retroviral class of drugs is a potent and specific HIV protease inhibitor used for the treatment of HIV infection is administered either orally or intravenously; while inhibiting the HIV viral protease enzyme which prevents cleavage of the gag-pol polyprotein which results in noninfectious, immature viral particles. Its dose range lies between 100mg to 400mg. Risks involved here are risk of fat redistribution, hemolytic anemia, hyperglycemia, hyperbilirubinemia, immune reconstitution syndrome if used in combination w/ other antiretroviral drugs. Indinavir was docked with proteases of SARS-COV-2 interesting facts about binding interaction were revealed. The docking of Indinavir showed a binding affinity score of -10.4 kcal/mol (Figure 8). Hydrogen bond was observed in Lys 5 and Arg 4. The oxygen bond interaction with Arg 4 of the B chain was also encountered. It also showed pi-alkyl interactions with Ile 286 and Lys 5 residues. When Indinavir was docked with COVID-19 proteases, two conventional hydrogen bonds were obtained. Arg 4 and Lys 5 of chain B formed hydrogen bonds, yielding a bond length of 3.22 and 3.25 respectively. Many hydrophobic interactions were encountered, showing a uniform involvement of residues present on both chains A and chain B respectively. These interactions were observed at Arg 4, Phe 3, Glu 288, Ser 284, Thr 285, Ile 286, Gln 127 of chain A and Gln 127, Tyr 126, Ile 286, Glu 288, Thr 285, Phe 291, Ser 284 of chain B. Comparatively Ligplot has generated a greater number of hydrophobic interactions between Indinavir and proteases.
- Zafirlukast belonging to the class of Anti-inflammatory, Anti-asthmatic oral leukotriene receptor antagonist (LTRA) for the maintenance treatment of asthma, binds to Cysteinyl leukotriene receptor 1 alters the cellular activity associated with the inflammatory process with a dose range from 10mg to 20mg. May cause eosinophilia, eosinophilic pneumonia, or vasculitis consistent with the emergence of Churg-Strauss syndrome Behavioral changes reported with therapy. Concomitant use with warfarin can result in a clinically significant increase in INR. Not for use in the treatment of acute asthma attacks, including status asthmaticus. Zafirlukast was docked against the proteases of SARS-COV-2, which gave an affinity score of -10.2 kcal/mol. It gives strong interaction with the proteases with its binding pocket between chain A and B (Figure 9). Thus, this interaction results in three conventional hydrogen bonds between oxygen and Arg 4 of chain A and Lys 5 of chain B respectively. Similarly, three carbon-hydrogen bonds were observed in association with Lys 5 and Lys 137. Four charged interactions were also encountered with Arg 131, Asp 289, Lys 5 of B chain and Arg 4 of A chain. Zafirlukast when docked against COVID-19 Proteases formed a conventional hydrogen bond with Lys 5 of chain B with a bond length of 2.92. This hydrogen bond was formed between the oxygen of the carbonyl group of ligand and nitrogen of lysine. Many hydrophobic interactions were observed at Lys 5, Glu 290, Arg 4, Phe 3of chain A and Arg 4, Phe 3, Glu 288, Asp 289, Arg 131, Lys 137, Try 126 and Gln 127 of B chain. As compared with another visualizer, Ligplot has revealed that Zafirlukast tends to show more tight interactions with a binding pocket region at the B chain of Covid-19 proteases.
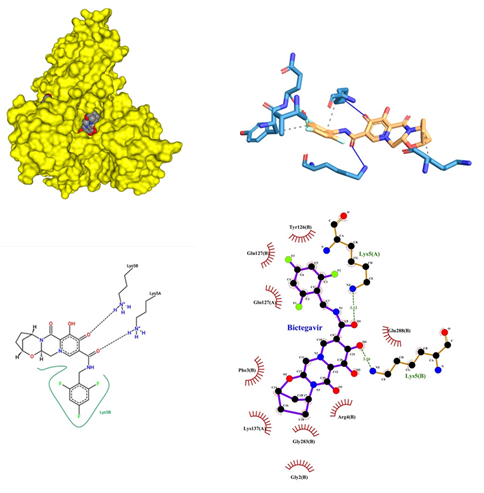
Figure 3: COVID-19 main proteases containing A and B chain.
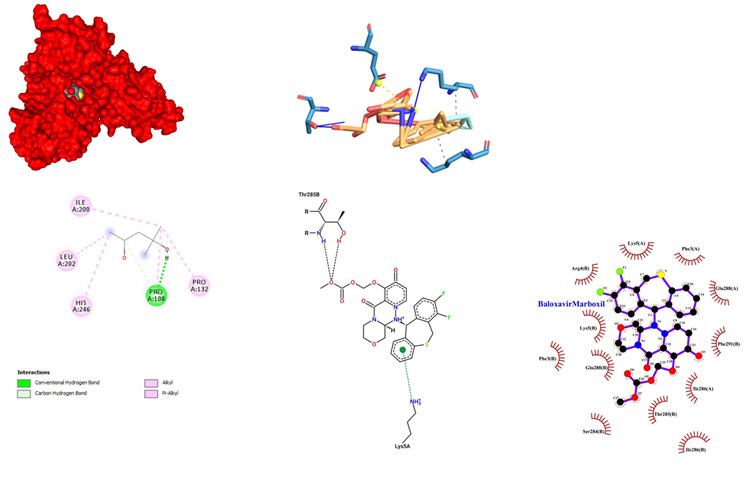
Figure 4: Baloxavir Marboxil docked in COVid-19 main protease (PDB ID 1Q2W) with (a) Best binding mode in the pocket of protein (with ligand as multi coloured space filled), (b) Amino acid residues involved in interaction (with ligand as orange stick), (c) Binding interaction of Baloxavir Marboxil with amino acid with hydrogen bond (green dash line), (d) 2D structure of Protein ligand interaction at the binding pocket.

Figure 5: Bictegravir docked in COVid-19 main protease (PDB ID 1Q2W) with (a) Best binding mode in the pocket of protein (with ligand as multi coloured space filled), (b) Amino acid residues involved in interaction (with ligand as orange stick), (c) Binding interaction of Bictegravir with amino acid with hydrogen bond (green dash line) and halogen bond (blue dash line), (d) 2D structure of Protein ligand interaction at the binding pocket.
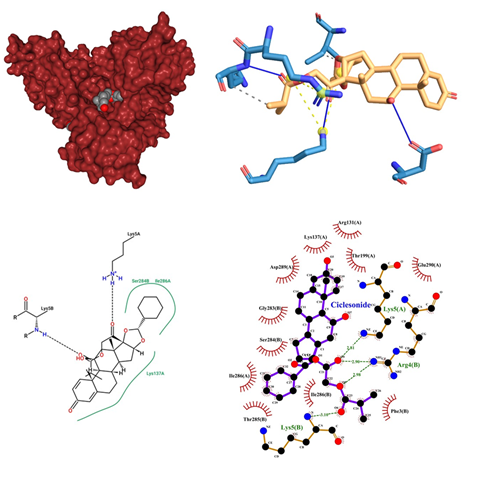
Figure 6: Ciclesonide docked in COVid-19 main protease (PDB ID 1Q2W) with (a) Best binding mode in the pocket of protein (with ligand as multi coloured space filled), (b) Amino acid residues involved in interaction (with ligand as orange stick), (c) Binding interaction of Ciclesonide with amino acid with hydrogen bond (green dash line) and alky bond (pink dash line), (d) 2D structure of Protein ligand interaction at the binding pocket.
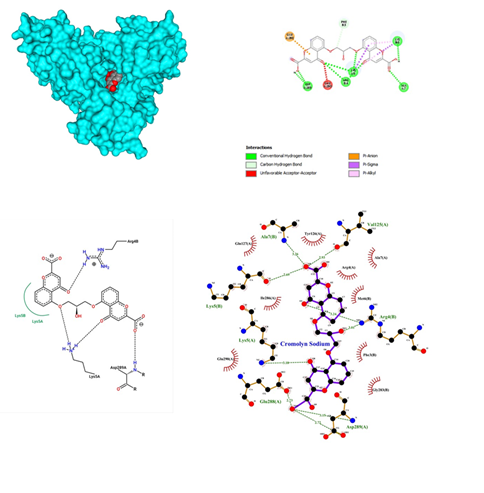
Figure 7: Cromolyn Sodium docked in COVid-19 main protease (PDB ID 1Q2W) with (a) Best binding mode in the pocket of protein (with ligand as multi coloured space filled), (b) Amino acid residues involved in interaction (with ligand as orange stick), (c) Binding interaction of Cromolyn Sodium with amino acid with hydrogen bond (green dash line), (d) 2D structure of Protein ligand interaction at the binding pocket.
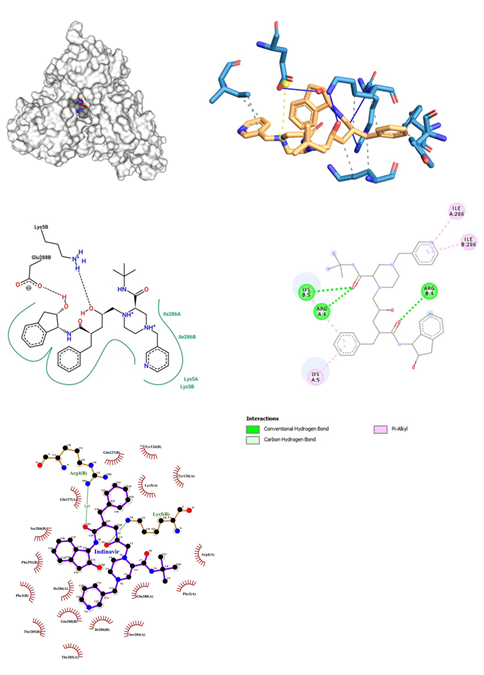
Figure 8: Indinavir docked in COVid-19 main protease (PDB ID 1Q2W) with (a) Best binding mode in the pocket of protein (with ligand as multi coloured space filled), (b) Amino acid residues involved in interaction (with ligand as orange stick), (c) Binding interaction of Indinavir with amino acid with hydrogen bond (green dash line) and alkyl (pink dash line), (d) 2D structure of Protein ligand interaction at the binding pocket.
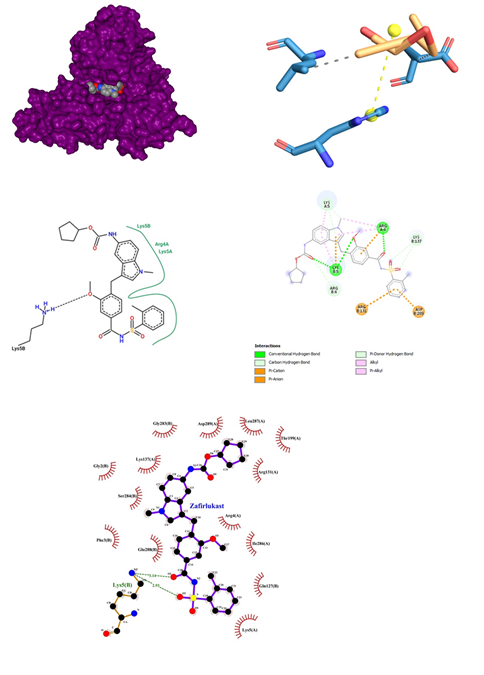
Figure 9: Zafirlukast docked in COVid-19 main protease (PDB ID 1Q2W) with (a) Best binding mode in the pocket of protein (with ligand as multi coloured space filled), (b) Amino acid residues involved in interaction (with ligand as orange stick), (c) Binding interaction of Zafirlukast with amino acid with hydrogen bond (green dash line), (d) 2D structure of Protein ligand interaction at the binding pocket.
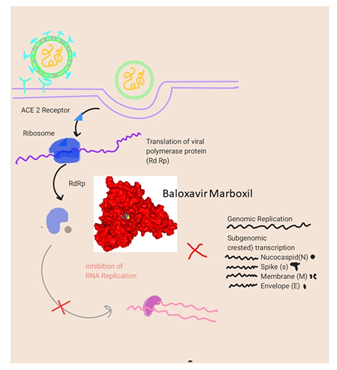
Figure 10: Mode of the mechanism of Baloxavir Marboxil when docked in COVID-19 proteases yielded 12 hydrophobic interactions.
Table no. 2 also predicts that Anti-asthma drugs like Zafirlukast, cromolyn sodium could be a better choice based on binding affinity, its mode of mechanism and its ability to be used as an antiviral agent, thereby trapping SARS-COV- 2 in the throat itself. This class of inhalers that could be used as antidotes before being infected subjected to further study findings. Inhalers could be the ideal choice of interest for further investigation. Now we will discuss In-silico modelling.
4. Discussion
The drugs that showed better binding affinity against the proteases of SARS-CoV-2 virus, belonged to the class of drugs like Antiviral drug B, Anti-Retroviral class, Anti-inflammatory and Anti-asthmatic where 3 drugs out of 6 drugs belonged to the anti-asthmatic class of drugs. The side-effects of drugs like Baloxavir Marboxil, Bictegravir, and Indinavir, with severe cross over effect and its inefficacy in fatal conditions as described in drugs description, inferred that asthmatic drugs seem to be more convenient, as compared to other drugs due to its less possibility of cross-over effect which could be a potential candidate for our expeditions to contain virus in the respiratory tract itself. This Molecular docking of 110 USFDA approved drugs, decreased the gap of understanding on how drugs are interacting with the SARS-COV-2 virus and also enlighten us with the key evidence that not just anti-inflammatory drugs are the target class of drugs but the use of asthmatic drugs like Cromolyn Sodium, Ciclesonide and Zafirlukast cannot be ruled out. The pandemic situation also made us compare the cost of these drugs, as when there is the treatment of mass in developing countries the major setback happens when the cost of drugs is higher than living. Considering this fact, the cost of shortlisted drugs played a significant role in this study. Asthmatic drugs like Cromolyn Sodium, Ciclesonide and Zafirlukast had the cost of approximately ranging from 1$ to 6$ as compared to antiviral drugs which are approximately ≤ 10$, hence this also provides evidence that anti-asthmatic can be considered. The pandemic situation has made us realise that; much study is needed on finding the right antiviral intervention is a key much more potent than vaccine once the damage of infection is done as vaccines are beneficial only in terms of creating anti-bodies than actually inducing a cidal effect on a virus which could be termed as virucidal. Much more investigations should be made on this new class of drugs known as virucidal rather than just antiviral.
5. Conclusion
Based on the discussion it is observed that out of 110 drugs, only 06 drugs having higher binding affinity were subjected to visualization; baloxavir marboxil, ciclesonide and indinavir has higher binding affinity than the rest of the drugs. Baloxavir Marboxil docked with SARS-COV-2 main proteases showed a significant binding with an affinity score of -10.6 kcal/mol. The interaction of Baloxavir Marboxil with the proteases showed a high-affinity interaction in chain A. This is further evidenced by hydrogen bonding between the oxygen of the carbonyl group of Baloxavir Marboxil with Pro 108. Some other Van der Waals interaction Baloxavir Marboxil with Pro 132, His 246, Leu 202 and Ile 200 has been observed. Ciclesonide docked in SARS-COV-2 main proteases shows a significant interaction in the central pocket near chain A and B with the binding affinity of -10.5 kcal/mol. The major interaction between Ciclesonide and the proteases is up-hold by hydrogen bonds between the oxygen group of ligands and Chain A Lys 5 and also with Arg 4 of chain B. An unfavourable acceptor interaction has also been observed at Glu 290. It also yielded a hydrogen bond connecting B chain Lys 5 with the oxygen of ligand (Ciclesonide) molecule. Some of the alkyl interactions of the aromatic ring and Ile 286 and Lys 137 have been encountered. Indinavir was docked with proteases of SARS-COV-2 interesting facts about binding interaction were revealed. The docking of Indinavir showed a binding affinity score of -10.4 kcal/mol. A hydrogen bond was observed in Lys 5 and Arg 4. The oxygen bond interaction with Arg 4 of the B chain was also encountered. It also showed pi-alkyl interactions with Ile 286 and Lys 5 residues. On the contrary, asthma drugs like Cromolyn sodium, Ciclesonide and Zafirlukast could be a better option as inhalers show fewer side effects while being cost-effective.
Acknowledgement
We are thankful to Guru Nanak Institute of Research and Development (GNIRD), a wing of our college Guru Nanak Khalsa College, Mumbai, for letting us work on this wonderful domain of Bioinformatics. Would also let our gratitude of thanks to Dr Narendra Chirmule for guiding us in this research work.
References
- Borba M, de Almeida Val F, Sampaio VS, Alexandre MA, Melo GC, et al. Chloroquine diphosphate in two different dosages as adjunctive therapy of hospitalized patients with the severe respiratory syndrome in the context of coronavirus (SARS-CoV-2) infection: Preliminary safety results of a randomized, double-blinded, phase IIb clinical trial (CloroCovid-19 Study). MedRxiv (2020).
- David M. COVID-19 (CORONAVIRUS): A Global Emergency Outbreak and its implications in India. Escors D, Ortego J, Enjuanes L.
- The membrane M protein of the transmissible gastroenteritis coronavirus binds to the internal core through the carboxy-terminus. InThe Nidoviruses. Springer, Boston, MA (2001).
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. Clinical characteristics of coronavirus disease 2019 in China. New England journal of medicine (2020).
- Jamwal A, Bhatnagar S, Sharma P. Coronavirus disease (COVID-19): Current literature and status in India (2019).
- Leung WK, To KF, Chan PK, Chan HL, Wu AK, Lee N, Yuen KY, Sung JJ. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003 Oct 1.
- Cai J, Xu J, Lin D, Xu L, Qu Z, Zhang Y, Zhang H, Jia R, Wang X, Ge Y, Xia A. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clinical Infectious Diseases. 2020 Feb 28
- Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology journal (2005).
- Shen K, Yang Y, Wang T, Zhao D, Jiang Y, Jin R, Zheng Y, Xu B, Xie Z, Lin L, Shang Y. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: experts’ consensus statement. World journal of pediatrics. 2020 Feb 7.
- J Alsaadi, E. A., & Jones, I. M. (2019). Membrane binding proteins of coronaviruses. Future Virology.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annual review of virology. 2016 Sep 29.
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virology journal. 2019 Dec.
- Masters PS. The molecular biology of coronaviruses. Adv. Virus Res. 66, 193–292 (2006).
- Klumperman J, Locker JK, Meijer A, Horzinek MC, Geuze HJ, Rottier PJ. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68(10), 6523–6534 (1994).
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol.
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4 (2012).
- Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. European journal of cell biology (1984).
- Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol (2013).
- McBride R, Van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses (2014).
- Chang KW, Sheng Y, Gombold JL. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology 269 (2000).
- Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. Virol (2010).
- Tseng YT, Wang SM, Huang KJ, Wang CT. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. J. Biomed. Sci 21 (2014).
- Lopez LA, Riffle AJ, Pike SL, Gardner D, Hogue BG. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol 82 (2008).
- Ujike M, Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses 7 (2015).
- Corse E, Machamer CE. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74 (2000).
- Gomez GN, Abrar F, Dodhia MP, Gonzalez FG, Nag A. SARS coronavirus protein nsp1 disrupts localization of Nup93 from the nuclear pore complex. Biochemistry and Cell Biology. 2019.
- Davies JP, Almasy KM, McDonald EF, Plate L. Comparative multiplexed interactomics of SARS-CoV-2 and homologous coronavirus non-structural proteins identifies unique and shared host-cell dependencies. bioRxiv. 2020 Jan 1.
- Saikatendu KS, Joseph JS, Subramanian V, Clayton T, Griffith M, Moy K, Velasquez J, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1 ″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005 Nov 1.
- Sakai Y, Kawachi K, Terada Y, Omori H, Matsuura Y, Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017 Oct 1;510:165-74.
- Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014 Aug 20.
- Lin CW, Tsai FJ, Wan L, Lai CC, Lin KH, Hsieh TH, Shiu SY, Li JY. Binding interaction of SARS coronavirus 3CLpro protease with vacuolar-H+ ATPase G1 subunit. FEBS letters. 2005 Nov 7.
- Protein Ligand Interaction Profiler Server, Available from: projects.biotec.tu-dresden.de/plip-web/plip
- Protein Plus Server, Available from:- proteins.plus
- Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life sciences. 2020 Feb 28:117477.
- Jokhakar PH, Kalaria R, Patel HK. In Silico Docking Studies of Antimalarial Drug Hydroxychloroquine to SARS-CoV Proteins: An Emerging Pandemic Worldwide.
- Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. Journal of Biological Chemistry. 2020 Sep 11;295(37):12910-34.
