Receiver Operating Characteristic and Recursive Linear Modeling for Gene Expression Analysis and Its Application to Alzheimer’s Disease Diagnosis by Peripheral Blood Mononuclear Cells
Article Information
Aibing Rao*
Shenzhen Luwei (Biomanifold) Biotechnology Limited 10th Floor, Clou Building B, Baoshen Road, Nanshan District, Shenzhen, PR China
*Corresponding authors: Aibing Rao, Shenzhen Luwei (Biomanifold) Biotechnology Limited 10th Floor, Clou Building B, Baoshen Road, Nanshan District, Shenzhen, PR China.
Received: 25 April 2024; Accepted: 02 May 2024; Published: 23 May 2024
Citation: Aibing Rao. Receiver Operating Characteristic and Recursive Linear Modeling for Gene Expression Analysis and Its Application to Alzheimer’s Disease Diagnosis by Peripheral Blood Mononuclear Cells. Journal of Biotechnology and Biomedicine. 7 (2024): 204-213.
Share at FacebookAbstract
Background and objectives: Microarray and RNA-Seq for gene expression analysis often generate expression matrix of very high dimension. In biomarker discovery study, a small set of important genes is the objective for data analysis and discovery. Innovative and effective gene discovery algorithm is always desirable. In this study we introduced a novel gene expression modeling algorithm, called Recursive Linear Modeling (RLM). By combining single-variate analysis and RLM for peripheral blood mononuclear cells (PBMC) gene expression analysis, we established and validated two prediction models for alzheimer’s disease (AD) and mild cognitive impairment (MCI) diagnosis.
Methods: Publicly available PBMC gene expression data sets for AD/MCI were used to develop and demonstrate the algorithm. By comparing the AD/MCI group to the healthy control (HC) group respectively, firstly, each gene was analyzed as a single-variate predictor using ROC (receiver operating characteristic), and a heuristic gene candidate set was selected from the top according to AUC (area under the curve) in the decreasing order. Secondly, for a given model size (number of genes in a model), the candidate set was searched by a recursive linear modeling procedure. At last, an optimal model size and the corresponding model was determined by the maximal R-square among all sizes.
Results: An AD prediction 30 gene model was established and validated with high specificity and sensitivity: SS18L2, ATP6V1G1, GIMAP7, OSBPL1A, C14orf166, UQCRH, USP3, STAT6, MFSD10, HELZ, FLT3, CBX7, PEPD, FGF7, ESD, REST, TM9SF3, ZNF264, LPAR1, CTGF, EML4, BTBD10, MED31, FCGRT, TAF12, SEC11C, FCER2, FASTKD2, RPS27A, RPS27. Its model building AUC is 0.98 and the validation AUC is 0.93; In parallel, an MCI prediction 23 gene model of similar performance was also established and validated: ULK1, UBL3, TPST2, EEF1A1, FAM21A, RAN, LCOR, NOD1, OSBPL1A, SARS, PAQR4, EGFL6, RPS23, SDHB, TFB1M, ZNF416, TRIP11, SEC22B, SELK, SDHC, SIPA1, ZSCAN21, OSGEPL1. Its model building AUC is 0.96 and the validation AUC is 0.88. The models may be used to develop accurate AD/MCI clinical diagnosis and early risk assessment.
Conclusions: A novel feature selection and model building method by combining single-variate analysis using ROC and recursive linear modeling was developed and its application to AD/MCI prediction based on PBMC expression data showed great accuracy. The method is very general and can be used to build models for other gene expression biomarker discovery studies.
Keywords
Alzheimer's disease; mild cognitive impairment; recursive linear modeling; gene expression; peripheral blood mononuclear cell
Article Details
Abbreviations:
AD: alzheimer’s disease; AUC: area under the curve; HC: healthy control; FPR: false positive rate; MCI: mild cognitive impairment; PBMC: peripheral blood mononuclear cell RLM: recursive linear modeling; ROC: receiver operating characteristic; TPR: true positive rate.
Introduction
Alzheimer’s Disease (AD) is a progressive neurodegenerative disease that mainly affects the elderly and seriously impairs their quality of life. In recent years, the number of people affected has rapidly increased, with over 10% of elderly people aged 65 or above suffering from AD. Due to the aging trend of society, AD has become one of the fastest-growing causes of death. It is estimated that by 2050, up to 100 million elderly people worldwide will be affected. In China, according to data from the National Health Commission, the prevalence of Alzheimer’s disease is 5.56%. There are approximately 15 million dementia patients among the elderly aged 60 and above, of which 10 million are AD. Mild Cognitive Impairment (MCI) is an intermediate state between normal aging and dementia, which involves one or more aspects such as memory, language, and judgment, leading to corresponding clinical symptoms, but daily abilities are not significantly affected. MCI may be caused by early AD. The symptoms of MCI may stabilize for several years or develop into AD or other types of dementia. In some cases, MCI may improve over time. At present, the diagnostic guidelines for AD recommend four aspects: 1. Medical history and clinical manifestations: early symptoms include decreased memory and lack of concentration; 2. Neuropsychological testing: using standard cognitive tests such as Mini Mental State Examination (MMSE); 3. Imaging examination: brain MRI or CT scan to exclude other causes; 4. Laboratory examination: Blood and urine tests exclude other causes. The secondary criteria involve recent immunological diagnostic methods, such as increased tau protein concentration in cerebrospinal fluid (CSF) The concentration of amyloid protein decreases, etc. Peripheral blood biomarkers also include tau protein and Amyloid protein. Traditional diagnosis of AD requires the condition to develop to an observable level, with invasive cerebrospinal fluid examination, peripheral blood tau protein, and The immuno-diagnostic method for amyloid protein has only recently begun, and its stability and accuracy still need to be verified. The diagnosis of MCI also includes four aspects: first, the patient complained of decreased memory, lack of concentration, and impact on daily life. 2. Physical examination and neurological assessment: exclude the possibility of other neurological diseases, such as Parkinson’s disease, Huntington’s disease, etc. 3. Cognitive assessment: Using standardized cognitive assessment tools to assess the cognitive function of patients. 4. Activity restriction assessment: Assess the patient’s daily living ability and degree of activity restriction. It can be seen that the diagnosis of MCI is more likely to be subjective.
Due to the advancement of microarray and NGS (next generation sequencing) technology, molecular diagnostic methods based on multigene expression analysis have been explored extensively for biomarker discovery. It can also provide new diagnostic methods that complement the above AD diagnostic criteria and provide more accurate early diagnostic tools for AD or for MCI.
Materials and Methods
Training and testing data sets The training data set GSE63061 and the testing data set GSE63060 were downloaded from the Gene Expression Omnibus (GEO). GSE63061 contains microarray data of 3 groups of PBMC samples from 135 (HC), 140 (AD) and 112 (MCI) subjects; GSE63060 contains microarray data of 3 groups of PBMC samples from 104 (HC), 145 (AD) and 80 (MCI) subjects. Both data sets were pre-processed as follows, at first, a normalization procedure was applied to each probe and then to each sample. The normalization is a linear map: (Q25, Q75) → (0, 1) where Q25, Q75 are the 25th and 75th percentile of a data vector; second, an average was taken with the normalized values of the probes mapped to the same gene and assigned to the gene; third, genes annotated by gencode.v22.annotation (https://www.encodeproject.org/files/gencode.v22.annotation/) as ”protein coding type” were used for the analysis. Moreover, genes missing in one of the data sets were omitted and therefore the training and the testing data sets contained 12235 common genes. In the following, subsets containing only HC and AD of both data sets were used for AD model building and validating while subsets containing only HC and MCI were used for AD model building and validating respectively.
The gene candidate sets determined by ROC: Given a training data set as defined in the above, for each gene, the receiver operating characteristic (ROC) method was applied to classify the disease group (either AD or MCI) and the healthy group (HC) respectively. A ROC curve was plotted with the false positive rate (FPR) as the horizontal axis and the true positive rate (TPR) as the vertical axis by running through a series of possible expression threshold of the gene. The series of threshold was obtained by binning the expression range with a fixed step size. At each threshold, label all samples below it as 0 and 1 otherwise, and then calculate (FPR, TPR) based on the sample truth, AD=1 or MCI=1, and HC=0. If a ROC curve for a gene is below the diagonal line, then 0-expression was used to replotted the curve, indicating that the gene is under-expressed. The AUC (area under the curve) was then calculated. It is worthwhile to note that an optimal cutoff is usually set at the position on the ROC curve which is the closest to the left-top corner with coordinate (0,1), where FPR = 0 and TPR=1, representing the perfect classification. Next sort the AUC in the decreasing order so that all genes were sorted according to their prediction powers. The sorted AUC for AD and MCI are plotted in (Figure 1). The ordered AUC trending curves show that the top 5% genes have remarkable prediction powers and hence were selected as candidates. The 95th percentiles of the AUC for AD and MCI is 0.63 and 0.62 respectively. By choosing genes with AUC greater than or equal to the 95th percentiles, we obtained 613 candidate genes for AD and 788 candidate genes for MCI. Note that there are 330 common genes in both AD and MCI candidate sets. By selecting the gene candidates, the gene searching universe for modeling is dramatically reduced.
Figure 1: Ordered single gene AUC in the decreasing order. The total number of genes is 12235 and the x-axis labels are only for displaying purpose. Both plots show that the top 5% genes have remarkable prediction powers and hence were selected as candidates for model building. AD: alzheimer’s disease; AUC: area under the curve; HC: healthy control; MCI: mild cognitive impairment.
Recursive linear modeling (RLM): A clinically practical gene expression assay typically contains a handful to tens of genes, hence a heuristic model size should be considered likewise. RLM takes a given model size S and searches the gene candidate space, denoted as G, to find an optimal linear model of size no more than S. G is an ordered gene list with decreasing AUCs calculated as in the above. In a typical linear regression model, along with each dependent variable there is a returned p value, which defines the statistic significance of the variable in the model fitting. A threshold p0 is used to omit genes with p > p0 and is typically set as 0.05. The genes with p ≤ p0 are kept. In more details, in the first round, RLM evenly partitions G into disjointed sublists of the equal size S. For each sublist, a linear model is built and the genes with p ≤ p0 are used to build another linear model, repeat it iteratively until all genes in the model have p ≤ p0. Take the union of the model genes by this iterative linear modeling method for all of the sublists, denoted as the new candidate gene set G, and repeat the above procedure recursively until the size of G is no more than S, i.e. |G| ≤ S.
Next an optimal model size is searched by running RLM through a series of model sizes. The optimal one has the highest average R2 among all model sizes. In the current implementation, the optimal model size was determined by searching model sizes from 10 to 60 for both AD and MCI models. This searching range was determined heuristically by considering the clinical diagnosis feasibility and the prediction power.
Data analysis and software RLM and plots were implemented in R scripts. The ROC analysis was based on R package ROCR.
Results
The AD linear model and its validation The RLM algorithm on the training subset of GSE63061 of the AD and the HC samples with 613 candidate genes gave rise to the 30 gene AD model, consisting of SS18L2, ATP6V1G1, GIMAP7, OSBPL1A, C14orf166, UQCRH, USP3, STAT6, MFSD10, HELZ, FLT3, CBX7, PEPD, FGF7, ESD, REST, TM9SF3, ZNF264, LPAR1, CTGF, EML4, BTBD10, MED31, FCGRT, TAF12, SEC11C, FCER2, FASTKD2, RPS27A, RPS27. The model coefficients are listed in Table 1. The sample AD scores was calculated as the weighted sum of the model gene expression values with the corresponding weights (estimates) shown in Table 1. The model building ROC using the AD score to predict sample groups (AD=1, HC=0) is presented in Figure 2. As shown in the figure, the model fits excellently with AUC = 0.98, the sensitivity (TPR) is 93%, the specificity (1-FPR) is 92% and the accuracy is 92%. Taking the testing subset of GSE63060 with the AD and the HC samples, the model is validated. The validating ROC is presented in Figure 3 which shows that AUC = 0.93, sensitivity = 82%, specificity = 87% and accuracy=84%.
|
varn |
pv |
estimate |
stderr |
tv |
rsq |
|
Intercept |
0 |
0.5103 |
0.0776 |
6.5795 |
0.6461 |
|
SS18L2 |
1.00E-04 |
0.36 |
0.0892 |
4.0374 |
0.6026 |
|
ATP6V1G1 |
2.00E-04 |
0.3281 |
0.0866 |
3.7877 |
0.6461 |
|
GIMAP7 |
1.00E-04 |
0.2138 |
0.0544 |
3.9297 |
0.6461 |
|
OSBPL1A |
2.00E-04 |
0.1789 |
0.0466 |
3.8394 |
0.6461 |
|
C14orf166 |
0.0091 |
0.1495 |
0.0569 |
2.6281 |
0.6461 |
|
UQCRH |
0.0256 |
0.1403 |
0.0624 |
2.2465 |
0.6461 |
|
USP3 |
6.00E-04 |
0.1323 |
0.038 |
3.4796 |
0.6461 |
|
STAT6 |
7.00E-04 |
0.1252 |
0.0363 |
3.4519 |
0.6461 |
|
MFSD10 |
0.0088 |
0.1064 |
0.0403 |
2.6414 |
0.6461 |
|
HELZ |
0.0063 |
0.0983 |
0.0357 |
2.7543 |
0.6461 |
|
FLT3 |
0 |
0.0978 |
0.0237 |
4.1323 |
0.6461 |
|
CBX7 |
0.0049 |
0.0903 |
0.0318 |
2.8416 |
0.6461 |
|
PEPD |
0.0494 |
0.0779 |
0.0394 |
1.9749 |
0.6461 |
|
FGF7 |
0.0049 |
0.0677 |
0.0238 |
2.839 |
0.6461 |
|
ESD |
0.0331 |
-0.0867 |
0.0405 |
-2.1433 |
0.6461 |
|
REST |
0.0017 |
-0.0876 |
0.0276 |
-3.1718 |
0.6461 |
|
TM9SF3 |
0.0318 |
-0.0885 |
0.041 |
-2.1598 |
0.6461 |
|
ZNF264 |
0.0042 |
-0.089 |
0.0308 |
-2.8864 |
0.6461 |
|
LPAR1 |
0.0138 |
-0.0937 |
0.0378 |
-2.4804 |
0.6461 |
|
CTGF |
0.0073 |
-0.0945 |
0.0349 |
-2.7059 |
0.6461 |
|
EML4 |
0.0097 |
-0.1102 |
0.0423 |
-2.6076 |
0.6461 |
|
BTBD10 |
0.0184 |
-0.1128 |
0.0475 |
-2.3737 |
0.6461 |
|
MED31 |
0.0076 |
-0.1153 |
0.0429 |
-2.6901 |
0.6461 |
|
FCGRT |
0.0084 |
-0.1261 |
0.0475 |
-2.656 |
0.6461 |
|
TAF12 |
7.00E-04 |
-0.1269 |
0.0368 |
-3.4433 |
0.6461 |
|
SEC11C |
0.017 |
-0.1374 |
0.0572 |
-2.4029 |
0.6461 |
|
FCER2 |
0 |
-0.147 |
0.0316 |
-4.6491 |
0.6461 |
|
FASTKD2 |
0 |
-0.1604 |
0.038 |
-4.2231 |
0.6461 |
|
RPS27A |
0 |
-0.3011 |
0.0633 |
-4.7583 |
0.6461 |
|
RPS27 |
0 |
-0.3301 |
0.0765 |
-4.3146 |
0.6461 |
Table 1: Coefficients of the 30 gene AD linear model ordered decreasingly by estimate. varn: variable; pv: p value; estimate: weight; stderr: standard error; tv: t value; rsq: R2
Figure 2: The model building ROC of the 30 gene AD model derived from the RLM procedure. AUC = 0.98, at the optimal cutoff (=0.5026) position, the sensitivity (TPR) is 93% and the specificity (1-FPR) is 92%. Therefore, the 30 gene AD model is an excellent fit to the training data. AD: alzheimer’s disease; AUC: area under the curve; HC: healthy control; MCI: mild cognitive impairment. RLM: recursive linear modeling; ROC: receiver operating characteristic.
Figure 3: The validating ROC of the 30 gene AD model tested on the subset of GSE63060 with the AD and the HC samples. It shows that AUC = 0.93, sensitivity = 82% and specificity = 87%. Therefore the 30 gene AD model is validated. AD: alzheimer’s disease; AUC: area under the curve; HC: healthy control; MCI: mild cognitive impairment. ROC: receiver operating characteristic.
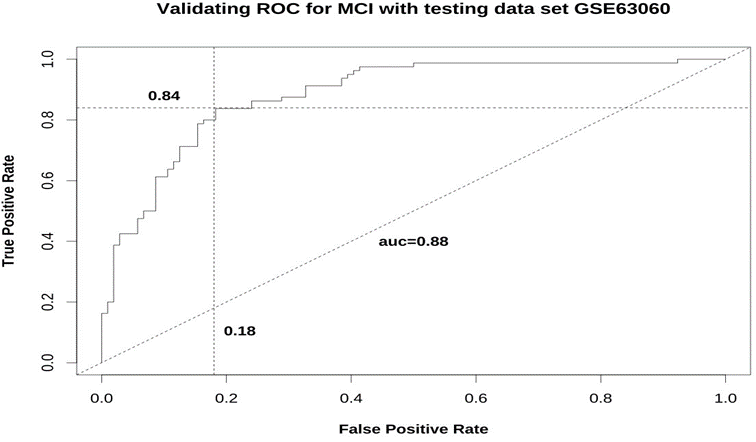
The MCI linear model and its validation The RLM algorithm on the training subset of GSE63061 of the MCI and the HC samples with 788 candidate genes gave rise to the 23 gene MCI model consisting of ULK1, UBL3, TPST2, EEF1A1, FAM21A, RAN, LCOR, NOD1, OS- BPL1A, SARS, PAQR4, EGFL6, RPS23, SDHB, TFB1M, ZNF416, TRIP11, SEC22B, SELK, SDHC, SIPA1, ZSCAN21, OSGEPL1. The model coefficients are listed in Table 2. The sample MCI scores were calculated as the weighted sum of the model gene expression values with the corresponding weights (estimates) shown in Table 2. The ROC using the MCI score to predict sample groups (MCI=1, HC=0) is presented in Figure 4. Again, the model fits greatly with AUC = 0.96, the sensitivity (TPR) is 88%, the specificity (1-FPR) is 90%, the accuracy is 89%. Taking the testing subset GSE63060 of the MCI and the HC samples, the model is validated. The validating ROC is presented in Figure 5 which shows that AUC = 0.88, sensitivity = 84%, specificity = 82%, and accuracy=83%.
|
varn |
pv |
estimate |
stderr |
tv |
rsq |
|
Intercept |
0.0126 |
0.2843 |
0.113 |
2.5165 |
0.5849 |
|
ULK1 |
1.00E-04 |
0.2107 |
0.0533 |
3.9522 |
0.542 |
|
UBL3 |
0.003 |
0.1998 |
0.0665 |
3.0044 |
0.5849 |
|
TPST2 |
0 |
0.1981 |
0.0411 |
4.8205 |
0.5849 |
|
EEF1A1 |
0.0117 |
0.1844 |
0.0726 |
2.5418 |
0.5849 |
|
FAM21A |
0 |
0.1707 |
0.0387 |
4.4087 |
0.5849 |
|
RAN |
0.0067 |
0.1599 |
0.0585 |
2.7359 |
0.5849 |
|
LCOR |
0 |
0.1571 |
0.0368 |
4.2645 |
0.5849 |
|
NOD1 |
2.00E-04 |
0.1428 |
0.0378 |
3.7768 |
0.5849 |
|
OSBPL1A |
0.0093 |
0.1338 |
0.051 |
2.623 |
0.5849 |
|
SARS |
0.0036 |
0.1171 |
0.0399 |
2.9396 |
0.5849 |
|
PAQR4 |
0.0032 |
0.1082 |
0.0362 |
2.9848 |
0.5849 |
|
EGFL6 |
7.00E-04 |
0.1017 |
0.0295 |
3.4518 |
0.5849 |
|
RPS23 |
0.0077 |
-0.0835 |
0.031 |
-2.6906 |
0.5849 |
|
SDHB |
0.0014 |
-0.1034 |
0.032 |
-3.2364 |
0.5849 |
|
TFB1M |
0.0017 |
-0.1048 |
0.0329 |
-3.1846 |
0.5849 |
|
ZNF416 |
0.0121 |
-0.1077 |
0.0426 |
-2.5297 |
0.5849 |
|
TRIP11 |
0.0029 |
-0.1155 |
0.0383 |
-3.011 |
0.5849 |
|
SEC22B |
7.00E-04 |
-0.1218 |
0.0354 |
-3.4373 |
0.5849 |
|
SELK |
0.0016 |
-0.122 |
0.0382 |
-3.1939 |
0.5849 |
|
SDHC |
7.00E-04 |
-0.1615 |
0.0467 |
-3.457 |
0.5849 |
|
SIPA1 |
0.0228 |
-0.171 |
0.0746 |
-2.292 |
0.5849 |
|
ZSCAN21 |
0 |
-0.176 |
0.0399 |
-4.4144 |
0.5849 |
|
OSGEPL1 |
3.00E-04 |
-0.1848 |
0.0498 |
-3.711 |
0.5849 |
Table 2: Coefficients of the 23 gene MCI linear model ordered decreasingly by estimate. varn: variable; pv: p value; estimate: weight; stderr: standard error; tv: t value; rsq: R2
Figure 4: The model building ROC of the 23 gene MCI model derived from the RLM procedure. It shows that AUC = 0.96, with MCI score cutoff = 0.4910, the sensitivity (TPR) is 88% and the specificity (1-FPR) is 90%. AD: alzheimer’s disease; AUC: area under the curve; HC: healthy control; MCI: mild cognitive impairment. RLM: recursive linear modeling; ROC: receiver operating characteristic.
Figure 5: The validating ROC of the 23 gene MCI model tested on the subset of GSE63060 with the MCI and the HC samples. It shows that AUC = 0.88, sensitivity = 84% and specificity = 82%. Therefore the 23 gene MCI model is validated. AD: alzheimer’s disease; AUC: area under the curve; HC: healthy control; MCI: mild cognitive impairment. ROC: receiver operating characteristic.
Comparison with other published methods Lunnon K, et. al. [1] presented a 48-gene classifier with an accuracy of 75% based on PBMC gene expression. Sanjana S, et. al [2] reported that the multi-tissue health aging signature has an AUC in 0.66-0.73 when being applied to PBMC expression data. Cheng L, et al. [3] published an exosome microRNA-based AD signature with sensitivity 87% and specificity 77%. Wang H, et. al. [5] used differential expression analysis and protein-protein interaction analysis to find an 8 gene signature: RPS17, RPL26, RPS3A, RPS25, EEF1B2, COX7C, HINT1, SNRPG. The AUCs of linear regressions with the 8 gene signature are: GSE63060 (AD: 0.88, MCI: 0.84) and GSE63061 (AD: 0.77, MCI: 0.80). GSE63060 was used as one of the training data sets in their analysis and hence has a better AUC. Nevertheless, the RLM models for AD (AUC: 0.93-0.98) and MCI (AUC: 0.88-0.96) have shown better AUCs. Interestingly, our AD and MCI models share no common gene with the 8 gene signature. On the other hand, in a review by Budelier MM, et. al. [4] on blood-based protein biomarkers with various assay techniques, among about 39 studies, the reported AUC ranged from 0.74 to 0.98, with an average of 0.87. Therefore, the 30 gene AD model and the 23 gene MCI model have a compatible accuracy comparing to the plasma protein biomarkers.
Discussion
It is not surprising to find numerous literatures on the roles of the AD and MCI model genes on neurodegenerative and intellectual disability diseases. Some of the model genes with high weight (estimate) magnitudes (at the top for the positive ones or at the bottom for the negative ones) were reviewed as follows. OSBPL1A (oxysterol binding protein like 1a) is the only shared gene in the AD and in the MCI model. OSBPL1A stabilizes GTP-bound RAB7A on late endosomes/lysosomes and alters functional properties of late endocytic compartments via its interaction with RAB7A, while RAB7A enhances tau secretion linked to the propagation of tau pathology [6]. OSBPL1A was also included in a promising gene signature predicting behavior changes of attention-deficit/hyperactivity disorder (ADHD) [7]. Now from the AD model shown in Table 1, the top gene with the highest positive weight is SS18L2 (SS18-Like protein 2) which is homologous to SS18. SS18 is a component of SWI/SNF (switch/sucrose nonfermenting) chromatin remodeling subcomplex. There have been a lot of researches on SWI/SNF complex and neurodevelopmental disorders or intellectual disability [8, 9]. Next on the list is ATP6V1G1 (ATPase H+ transporting V1 subunit G1), which is a member of vacuolar-type ATPases (V- ATPases). V-ATPases and other types of ATPases have important roles in neurodegenerative diseases [10, 11, 12]. On the opposite negative weight side, the bottom two rows on Table 1 are two genes, RPS27 and RPS27A, with weight magnitude greater than 0.30. Ribosomal protein RPS27 was shown to be over-expressed in glioma [13]. RPS27A encodes part of ubiquitin. The ubiquitin-proteasome system predominantly driving protein aggregation in the age-related diseases such as Parkinson’s disease [14]. RPS27A was also inferred to be a controller of microglia activation in triggering neurodegenerative diseases [15]. Moreover RPS27A was documented by MalaCards to be related to the neuronal intranuclear inclusion disease of which the cognitive impairment might be one of the symptoms. The next AD gene with the positive weight is GIMAP7, the GTPase domain of the immune associated nucleotide binding protein 7. GIMAP7 might be through the AMPK signal pathway. GIMAP7 suppresses AMPK signal pathway in lung cancer cells [16] and it is unclear whether it is true in AD, while AMPK was reviewed to have controversially preventive and proactive roles on AD in different studies [17]. The next gene with the negative weight is FASTKD2 (fas activated serine/threonine kinase domain 2), which has a structure containing mitochondrial targeting domain, multiple serine/ threonine kinase domains and an RNA-binding domain. FASTKD2 might be involved with human memory via three possible pathways [18]: first, the neuroprotective effect of FASTKD2 on memory might be through fas-mediated apoptosis; second, the findings of rare mutations of FASTKD2 leading to cytochrome c oxidase (mitochondrial respiratory chain) deficiency or inherited ataxias, suggesting its involvement with mitochondrial dysfunction and closely related oxidative stress pathways which are strongly related to neurodegeneration in aging and disease; at last, FASTKD2 has a proinflammatory role while inflammation plays central roles in compensating cellular stress induced by amyloid-β deposition. Interestingly, another AD model gene UQCRC1 (human ubiquinol- cytochrome c reductase core protein 1) is also related to mitochondrial respiratory chain and engaged with neuronal apoptotic cell death [19]. There are several other genes with notable weights related to inflammation and immune system: USP3 deubiquitinates and stabilizes ASC [20] (apoptosis associated speck like protein containing a caspase recruitment domain), the adaptor for inflammasome activation, which was shown to be highly related to AD [21, 22]; STAT6 was implicated in several immunity-related pathological pathways [23] and was demonstrated to activate neural stem cell proliferation and neurogenesis upon amyloid-β42 aggregation with a zebrafish model [24]; FCER2 (Fcε receptor II) regulates immunoglobulin E (IgE) production and plays essential roles in the differentiation of B cells, while B cell depletion was shown to reverse AD progression [25, 26]; FCGRT (Fcγ receptor and transporter) encodes the heavy chain of neonatal Fc receptor (FcRn). FcRn binds to the Fc portion of IgGs, protects IgGs from degradation, facilitates IgG transport, and potentiates IgG related cellular immune responses. IgG was demonstrated to be an aging factor [27], FcRn promotes the development and progression of diseases of the nervous system [28], and a study demonstrated that FCGRT was elevated in the midbrain from schizophrenia patients with high inflammation [29], therefore FcRn and IgG may play important roles in the AD development.
Some genes of the MCI model are reviewed next. ULK1,UBL3,TPST2,EEF1A1 are the top 4 MCI genes with positive weights (Table 2) and OSGEPL1, ZSCAN21, SIPA1, SDHC are the bottom 4 with negative weights. ULK1 is a serine/threonine-protein kinase involved in autophagy in response to starvation and regulates autophagosome formation [30] where AMPK/mTOR serve as the accelerator/brake of ULK1 respectively under starvation or nutrient sufficiency conditions [31]. Autophagy were shown to play important roles in neurodegenerative diseases [32, 33]. UBL3 (ubiquitin-like 3) was demonstrated to interact with α-synuclein [34], which plays a critical role in the pathogenesis of PD and alike, and its aggregates perturb dopaminergic transmission and induce presynaptic and postsynaptic dysfunctions and cause neuroinflammation [35]. Interestingly, ZSCAN21, with a negative weight, was shown to stimulate α-synuclein gene SNCA transcription in neuronal cells. TRIM41 is an E3 ubiquitin ligase while TRIM17 decreases the TRIM41-mediated degradation of ZSCAN21. TPST2 (tyrosylprotein sulfotransferase 2) catalyzes tyrosine sulfation and was shown to contribute to long-term memory [37]. EEF1A1 (eukaryotic translation elongation factor 1 alpha 1) encodes an isoform of the alpha subunit of the elongation factor-1 complex, which is responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome. A study showed that EEF1A1 participates neuroinflammation in PD by regulating the inflammation delaying gene GDF15, STC1, MT1E, MT1X, GPNMB, VIP, A2M and the accelerating gene IL-6, CCL5 [38]. OSGEPL1 (o-sialoglycoprotein endopeptidase like 1) is required for t6A37 modification in mitochondrial tRNA [39] and mitochondrial stress was demonstrated to be highly related to MCI [40]. SIPA1 (signal-induced proliferation-associated protein 1) is a mitogen induced GTPase activating protein (GAP) and activates RAP1, a RAS family member of small GTPases. SIPA1 and RAP1 signaling plays the critical role in T cell β-selection checkpoint, namely the transition from CD4/CD8 double-negative (DN) to double-positive (DP) stage, and is crucial for T cell normal development [41, 42]. RAP1 signaling is also related to calcium signaling [43]. The association of SIPA1 expression on MCI might be through these two pathways. SDHC and SDHB are MCI model genes with negative weights. Succinate dehydrogenase (SDH) complex has 4 subunits SDHA/B/C/D and SDH involves with multiple neurodegenerative diseases [44].
Conclusions
A novel feature selection and model building method was proposed for gene expression analysis using ROC and RLM, its application to AD/MCI prediction based on public PBMC expression data set has given rise to a 30-gene AD prediction model and a 23-gene MCI prediction model, which were validated with independent data sets. The corresponding model building AUC for AD and MCI is 0.98 and 0.96, while the validatingAUC is 0.93 and 0.88 respectively, which are superior to other published results. Literature reviews confirmed that most of model genes were demonstrated to be highly relevant, although some other novel genes might be worthwhile for further investigation. The method is very general and can be applicaple to build models for any other gene expression biomarker discovery studies.
Declaration
Conflict of Interest Aibing Rao is a co-founder of Shenzhen Luwei (Biomanifold) Biotech- nology Limited, Shenzhen, China.
References
- Lunnon K, et al. A blood gene expression marker of early Alzheimer’s disease. J Alzheimers Dis 33 (2013): 737-53.
- Sanjana S, et al. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biology 16 (2015): 185.
- Cheng L, et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry (2014).
- Budelier MM, Bateman RJ. Biomarkers of Alzheimer Disease. J Appl Lab Med 5 (2020): 194-208.
- Wang H, Han X, Gao S. Identification of potential biomarkers for pathogenesis of Alzheimer’s disease. Hereditas 158 (2021): 23.
- Rodriguez L, et al. Rab7A regulates tau secretion. J Neurochem 141 (2017): 592-605.
- Suresh P, et. al. Evaluating the Neuroimaging-Genetic Prediction of Symptom Changes in Individuals with ADHD. Annu Int Conf IEEE Eng Med Biol Soc 2021 (2021): 1950-1956.
- Valencia AM, et al. Landscape of mSWI/SNF chromatin remodeling complex perturbations in neurodevelopmental disorders. Nat Genet. 55 (2023): 1400–1412.
- Santen GW, et al. SWI/SNF complex in disorder: SWItching from malignancies to intellectual disability. Epigenetics. 7 (2012): 1219-1224.
- Song Q, et al. The emerging roles of vacuolar-type ATPase-dependent Lysosomal acid- ification in neurodegenerative diseases. Transl Neurodegener 9 (2020): 17.
- Ebanks B, et al. ATP synthase and Alzheimer’s disease: putting a spin on the mitochon- drial hypothesis. Aging (Albany NY) 12 (2020): 16647-16662.
- Zhou Z, et al. Downregulation of ATP6V1A involved in alzheimer’s disease via synaptic vesicle cycle, phagosome, and oxidative phosphorylation. Oxid Med Cell Longev 2021 (2021): 5555634.
- Feldheim J, et al. Protein S27/metallopanstimulin-1 (RPS27) in glioma—a new disease biomarker? Cancers 12 (2020): 1085.
- Tiwari S, et al. UBA52 Is crucial in HSP90 ubiquitylation and neurodegenerative signaling during early phase of Parkinson’s disease. Cells 11 (2022): 3770.
- Khayer N, et al. Rps27a might act as a controller of microglia activation in triggering neurodegenerative diseases. PLoS One 15 (2020): e0239219.
- Cui L, et al. GIMAP7 inhibits epithelial-mesenchymal transition and glycolysis in lung adenocarcinoma cells via regulating the Smo/AMPK signaling pathway. Thorac Cancer 15 (2024): 286-298.
- Assefa BT, et al. The bewildering effect of AMPK activators in alzheimer’s disease: review of the current evidence. Biomed Res Int 2020 (2020): 9895121.
- Ramanan VK, et al. FASTKD2 and human memory: functional pathways and prospects for novel therapeutic target development for Alzheimer’s disease and age-associated memory decline. Pharmacogenomics 16 (2015): 429-432.
- Hung YC, et al. UQCRC1 engages cytochrome c for neuronal apoptotic cell death. Cell Rep 36 (2021): 109729.
- Zhuang W, et al. USP3 deubiquitinates and stabilizes the adapter protein ASC to regulate inflammasome activation. Cell Mol Immunol 19 (2022): 1141–1152.
- Venegas C, et al. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 552 (2017): 355–361.
- Scott XO, et al. The inflammasome adaptor protein ASC in mild cognitive impairment and Alzheimer’s disease. Int J Mol Sci 21 (2020): 4674.
- Karpathiou G, et al. STAT6: A review of a signaling pathway implicated in various diseases with a special emphasis in its usefulness in pathology. Pathol Res Pract 223 (2021): 153477.
- Bhattarai P, et al. IL4/STAT6 signaling activates neural stem cell proliferation and neurogenesis upon amyloid-β42 aggregation in adult zebrafish brain. Cell Rep 17 (2016): 941-948.
- Kim K, et al. Therapeutic B-cell depletion reverses progression of Alzheimer’s disease. Nat Commun 12 (2021): 2185.
- Plantone D, et al. B lymphocytes in Alzheimer’s diseasea comprehensive review. J Alzheimers Dis 88 (2022): 1241-1262.
- Yu L, et al. IgG is an aging factor that drives adipose tissue fibrosis and metabolic decline. Cell Metabolism 36 (2024): 793-807.
- Pyzik M, et al. The therapeutic age of the neonatal Fc receptor. Nat Rev Immunol 23 (2023): 415-432.
- Petty A, et al. Increased levels of a pro-inflammatory IgG receptor in the midbrain of people with schizophrenia. J Neuroinflammation 19 (2022): 188.
- Zachari M, et al. The mammalian ULK1 complex and autophagy initiation. Essays Biochem 61 (2017): 585-596.
- Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13 (2011): 132-141.
- Nagayach A, et al. Autophagy in neural stem cells and glia for brain health and diseases. Neural Regen Res 19 (2024): 729-736.
- Griffey CJ, et al. Macroautophagy in CNS health and disease. Nat Rev Neurosci 23 (2022): 411–427.
- Chen B, et al. UBL3 Interacts with Alpha-synuclein in cells and the interaction Is down- regulated by the EGFR pathway inhibitor Osimertinib. Biomedicines 11 (2023): 1685.
- Calabresi P, et al. Alpha-synuclein in Parkinson’s disease and other synucleinopathies: from overt neurodegeneration back to early synaptic dysfunction. Cell Death Dis 14 (2023): 176.
- Lassot I, et al. The E3 ubiquitin ligases TRIM17 and TRIM41 modulate α-synuclein expression by regulating ZSCAN21. Cell Rep 25 (2018): 2484-2496.e9.
- Sahoo B, et al. Sulfotransferase activity contributes to long-term potentiation and long- term memory. Learn Mem 29 (2022): 155-159.
- Aisha Z, et al. EEF1A1 is involved the regulating neuroinflammatory processes in Parkin- son’s disease. J Integr Neurosci 22 (2023): 122.
- Zhou JB, et al. Molecular basis for t6A modification in human mitochondria. J Nucleic Acids Res 48 (2020): 3181-3194.
- Kim KM, et al. Mitochondrial RNA in Alzheimer’s Disease Circulating Extracellular Vesicles. Front Cell Dev Biol 8 (2020): 581882.
- Minato N, et al. Spa-1 (Sipa1) and Rap signaling in leukemia and cancer metastasis. Cancer Science 100 (2009): 17-23.
- Horitani S, et al. The critical role of Rap1-GAPs Rasa3 and Sipa1 in T cells for pulmonary transit and egress from the lymph nodes. Front Immunol 14 (2023): 1234747.
- Kosuru R, et al. Integration of Rap1 and Calcium Signaling. Int J Mol Sci 21 (2020): 1616.
- Farshbaf MJ, et al. Succinate dehydrogenase: Prospect for neurodegenerative diseases, Mitochondrion 42 (2018): 77-83.