Identification of a Papain-Like Protease Inhibitor with Potential for Repurposing in Combination with an Mpro Protease Inhibitor for Treatment of SARS-CoV-2
Article Information
Jesus Campagna1, Barbara Jagodzinska1, Pablo Alvarez2, Constance Yeun4, Kathryn M Enquist1, Whitaker Cohn1, Pavla Fajtová3, Anthony J O’Donoghue3, Vaithilingaraja Arumugaswami4, Melody MH Li2, Robert Damoiseaux4,5,6,7 and Varghese John1*
1The Drug Discovery Lab, Department of Neurology, David Geffen School of Medicine, 710 Westwood Plaza, University of California Los Angeles, Los Angeles, CA 90095, USA
2Department of Microbiology, Immunology & Molecular Genetics, 615 Charles E. Young Drive, University of California Los Angeles, Los Angeles, CA 90095, USA
3Skaggs School of Pharmacy and Pharmaceutical Sciences, 9500 Gilman Drive, University of California San Diego, La Jolla, CA 92093, USA
4Department of Molecular and Medical Pharmacology, 650 Charles E. Young Drive, University of California Los Angeles, Los Angeles, CA 90095, USA
5Department of Bioengineering, University of California, Los Angeles, CA 90095, USA
6California NanoSystems Institute, University of California, Los Angeles, CA 90095, USA
7Jonsson Comprehensive Cancer Center, University of California, Los Angeles, CA 90095, USA
*Corresponding author: Varghese John, The Drug Discovery Lab, Department of Neurology, David Geffen School of Medicine, 710 Westwood Plaza, University of California Los Angeles, Los Angeles, CA 90095, USA.
Received: 01 December 2022; Accepted: 09 December 2022; Published: 17 December 2022
Citation:
Jesus Campagna, Barbara Jagodzinska, Pablo Alvarez, Constance Yuen, Kathryn M Enquist, Whitaker Cohn, Pavla Fajtová, Anthony J O’Donoghue, Vaithilingaraja Arumugaswami, Melody MH Li, Robert Damoiseaux and Varghese John. Identification of a Papain-Like Protease Inhibitor with Potential for Repurposing in Combination with an Mpro Protease Inhibitor for Treatment of SARS-CoV-2. Archives of Clinical and Biomedical Research 6 (2022): 993-1002.
Share at FacebookAbstract
SARS-CoV-2 requires two cysteine proteases for viral polypeptide processing to allow maturation and replication: the 3C-like protease also known as the Main protease (Mpro) and the papain-like protease (PLpro). In addition to its critical role in viral replication, PLpro removes post-translational modifications like ubiquitin and interferon-stimulated gene product 15 (ISG15) from host proteins through its deubiquitinase domain, leading to host immunosuppression and increased ability of the virus to evade the host antiviral immune response. Through screening of a custom clinical compound library, we identified eltrombopag (DDL-701), a thrombopoietin receptor agonist, as having PLpro inhibitory activity that is sustained in the presence of the Mpro inhibitor nirmatrelvir. DDL-701 also suppressed both the deubiquitinase and ISG15 cleavage activities of PLpro. In addition, DDL- 701 partially restored interferon-β induction – an element of the host immune response – in an in vitro model system. Further, modeling and docking studies suggest DDL-701 interacts with the active site region of the PLpro enzyme and pilot pharmacokinetic studies indicate it is brain permeable. DDL-701 is already FDA approved for treatment of thrombocytopenia and has previously been shown to achieve human plasma levels after oral dosing that is above the IC50 needed for it to exert its PLpro inhibitory activity in vivo. In addition, it has also been reported to have antiviral efficacy against SARS-CoV-2. DDL-701 thus represents an excellent drug candidate that can immediately be repurposed and undergo clinical evaluation as a PLpro inhibitor, that most effectively in the clinic may be used in a protease inhibitor cocktail with an Mpro inhibitor such as nirmatrelvir (Paxlovid) for the treatment of COVID-19.
Keywords
Antiviral Resistance; COVID-19; Drug Repurposing; Modeling; Mpro Inhibitor; Nirmatrelvir; Paxlovid; Rebound; PLpro Inhibitor; Protease Inhibitor Cocktail
Antiviral Resistance articles; COVID-19 articles; Drug Repurposing articles; Modeling articles; Mpro Inhibitor articles; Nirmatrelvir articles; Paxlovid articles; Rebound articles; PLpro Inhibitor articles; Protease Inhibitor Cocktail articles
Article Details
1. Introduction
Despite the development of vaccines, COVID-19 was the third-leading cause of death in the U.S. in 2021 [1] and new variants with the potential to evade vaccine protection have appeared, with more anticipated [2]. This will likely result in continued need for treatments that are effective post-infection. One strategy for development of antiviral medications to mitigate severe COVID-19 infections has been to target the two viral cysteine proteases, the 3C-like protease also known as the main protease (Mpro) and the papain-like protease (PLpro), that are essential for polypeptide processing during viral maturation and replication [3-5]. The coronavirus nonstructural protein 3 (nsp3) has multifunctional domains including one of the two viral proteases used for initial cleavage of the coronavirus polyprotein through its PLpro domain. In SARS-CoV, PLpro has been reported to mediate cleavage of interferon (IFN) stimulated gene product 15 (ISG15) from interferon regulatory factor 3 (IRF3), blocking its nuclear translocation, and reducing type-1 IFN responses leading to host immune suppression [6, 7]. Nsp3 has several domains outside of the PLpro domain including the nearby ubiquitin-like domain and macro domain [3]. It is thought that the ubiquitin-like domain works in concert with the PLpro domain to carry out its deubiquitinating and deISGylating activities, which disrupt the host innate immune response by removing important ISG15 molecules that help promote the type-1 IFN response [8]. Additionally, the macro domain is thought to antagonize the innate immune response by removing ADP-ribose, a post-translational modification that enhances interferon signaling [6]. Previous studies have identified nsp3 as a main immune antagonist in many coronaviruses including HCoV-NL63, SARS-CoV, MERS-CoV, and SARS-CoV-2 [6, 7, 9-12]. Interestingly, both SARS and MERS viruses contain SARS-unique domains in nsp3 that enhance the innate immune antagonism activity of the PLpro domain, which supports the merits of targeting the PLpro for therapeutic treatment against SARS-CoV-2 [3, 13]. With the progression of COVID-19, studies have found a strong correlation between innate immune deficiency and severe outcomes [14] to which the host immune antagonism activity of nsp3 likely contributes.
In initial efforts to identify a therapeutic for SARS-CoV-2 infection, Mpro inhibitors were targeted, leading to the development and FDA emergency use authorization of nirmatrelvir (Paxlovid) [15]. Mpro is a promising drug target because it is dissimilar from human proteases and plays a critical role during infection. Most research has so far only focused on developing single inhibitors for Mpro, and clinical data has shown that they successfully reduce hospitalization from COVID-19 [15]. Despite the benefits of nirmatrelvir treatment, there have been frequent reports of a ‘rebound’ effect [16, 17] that may be due, in part, to Mpro inhibition monotherapy not being completely effective in arresting SARS-CoV-2 replication. Recent studies suggest that the virus could develop resistance to nirmatrelvir through development of mutations in the protease Mpro, a phenomenon observed with many antiviral drugs [18, 19]. This has raised interest in the development of protease inhibitor cocktail therapy for SARS-CoV-2 [20-27] with PLpro and Mpro inhibitors to increase antiviral efficacy and reduce the risk of rebound, which is modeled on the use of protease cocktails for the treatment of HIV and hepatitis C. Use of a protease inhibitor cocktail could help reduce or prevent resistance by making it harder for the SARS-CoV-2 virus to evolve around multiple inhibitors.
Here, in pursuit of the objective of identifying a PLpro inhibitor that may be used in a protease inhibitor cocktail with an Mpro inhibitor, we screened a custom 58-compound library of pharmacologically active molecules, including FDA-approved drugs, for PLpro inhibition activity and identified the approved drug eltrombopag (DDL-701) that showed PLpro inhibitory activity with an IC50 that is achieved in human plasma after oral dosing. Importantly, the protease inhibition activities of both DDL-701 and nirmatrelvir are sustained when combined in vitro.
2. Methods
2.1 PLpro, Mpro, deubiquitinase, and ISG15 in vitro Assays
PLpro and PLpro deubiquitinase substrate kits each containing both the enzyme and substrate were purchased from BPS Bioscience (cat# 79995-2 & cat# 79996 respectively), and for the PLpro ISG15 substrate assay, rhodamine 110 was purchased from South Bay Bio (cat# SBB-PS0002), this kit also contains the PLpro enzyme and ISG15 substrate; each assay was performed following the manufacturer’s recommendations, in a 384-well plate format. For the Mpro assay, the Mpro enzyme was obtained from the O’Donoghue lab at UCSD, the fluorogenic substrate for SARS-CoV-2 Mpro was purchased from Vivitide (cat# SFP-3250-v). Briefly, in each assay, the enzyme was loaded into each well in the appropriate buffer, next the compounds were added into the well and incubated for 10-60 min at 37oC. The reaction was initiated by addition of substrate to each well and the fluorescent signal was read for 60 min at the appropriate excitation/emission. For the PLpro and PLpro deubiquitinase assays, DTT was added to the assay buffer to achieve 1mM DTT before the assay. A 10 mM stock of compounds was prepared in DMSO, this stock was diluted in assay buffer to achieve a 10X of the desire concentration in the well. Next, the PLpro and deubiquitinase substrates were diluted to 42 µM and 0.5 µM respectively in the assay buffer. Then, PLpro enzyme was diluted to 2.5ng/µL and 6ng/µL for the PLpro and deubiquitinase assays respectively in the assay buffer. The reaction was loading 2.5 µL of compounds to each well, then 10 µL of PLpro enzyme was added, follow by an incubation of 1 hours. The reaction was started by adding 12.5 µL of the substrate to each well, the reaction was monitored for 1h at excitation at 360 nm; emission at 460 nm. For the ISG15 substrate, PLpro enzyme at 0.1 nM was incubated with compounds for 1h and the reaction was initiated by adding ISG15-RH110 substrate at 500 nM, the reaction was monitored for 1-hour excitation at 485 nm; emission at 535 nm. For the scatterplot analysis for PLpro enzyme inhibitors from the custom clinical library compounds, treatment with compounds was done at 50 μM. For the PLpro enzyme inhibition in Figure 2A, DDL-701 was tested at 50 μM, DDL-750 (nirmatrelvir, MedChemExpress) at 0.1 μM and the combination of DDL-701/750 was tested at 50 μM/0.1 μM respectively. For the Mpro activity assay, 8 µL of Mpro at 5 µg/mL in assay buffer was loaded into each well, followed by 100 nL of the compounds at 50 μM and incubated for 10 min at 37oC. Following the incubation, 2 µL of substrate at 25 µM was added into the wells and the fluorescent signal (ex/em 380/455nm) was recorded for 1 h at 37oC. The sources and catalog numbers of the various drugs used in this study is shown in Supplementary Table S1.
2.2 Cloning of Expression and Reporter Constructs
The SARS CoV-2 papain-like protease (PLpro) domain of Nsp3 was cloned from a doxycycline-inducible piggyBac transposon vector (PB-TAC-ERP2, Addgene# 80478) containing the synthesized full-length Nsp3 from the Wuhan-Hu-1 SARS CoV 2 strain (Alvarez and Yao, unpublished). The PLpro domain (amino acids 745-1061) was cloned using PCR (Forward primer 5’-GTTTGCGGCCGCAAGAGAGGTGAGAACCATCAAG-3’, Reverse primer 5’- GGTGTCTAGATTAAGGTTTTATGGTTGTGGTATAG-3’) and inserted into myc-pcDNA3.1 using NotI and XbaI restriction sites. For the Renilla luciferase reporter construct, the Renilla luciferase gene was amplified from a Japanese encephalitis virus replicon containing a Renilla luciferase reporter gene using PCR (Forward primer 5’-GTTTAAGCTTGCCACCATGGCTTCCAAGGTGTAC-3’, Reverse primer 5’-TTTGCTCGAGTCACTGCTCGTTCTTCAGCAC-3’) and inserted into V5-pcDNA3.1 using HindIII and XhoI restriction sites, removing the V5 tag. An Addgene plasmid was used for the IFN-β promoter luciferase reporter (IFN-Beta_pGL3, #102597). To make the NF-κB signaling reporter, a gene block containing 5 tandem consensus NF-κB binding sites (Badr et al 2009; Ngo et al 2020), followed by the -55 to +19 region of the human IFN-β gene (UCSC Genome Browser) was synthesized by Integrated DNA Technologies (Alvarez and Yao, unpublished). The gene block was then moved into the pCRTM-Blunt II-TOPOTM vector using the Zero BluntTM TOPOTM PCR Cloning Kit (ThermoFisher) and using EcoRI and NheI to move the gene block into IFN-Beta_pGL3, replacing the IFN-β promoter. For SARS CoV-2 Mpro, the plasmid was provided by Rolf Hilgenfeld, University of Lubeck, Germany [28] and transformed into Escherichia coli strain BL21-Gold (DE3). The expression and purification of the protein has been described in detail previously [29].
2.3 IFN-β Induction and NFκB Signaling Luciferase Reporter Assays
To assess whether the compounds could rescue IFN-β induction, HEK293T cells were treated with compound by replacing the media with new media containing the compound at the desired concentration. The cells were then co-transfected with 150 ng of an IFN-β promoter-Firefly luciferase reporter plasmid, 10 ng of a Renilla luciferase expression plasmid, and 250 ng of either an empty myc-pcDNA3.1 vector or a plasmid expressing SARS-CoV-2 PLpro domain of Nsp3 (amino acids 745-1061), using the TransIT-mRNA Transfection Kit (Mirus bio). To induce IFN-β, 500 ng of poly I:C (InvivoGen) was added into the co-transfection mix. For the NFκB signaling assay, cells were co-transfected with 150 ng of an NFκB dependent-Firefly luciferase reporter plasmid wherein the luciferase gene expression was under control of 5 tandem NFκB binding sites followed by the -55 to +19 region of the human IFNB1 gene for correct spacing, 10 ng of a Renilla luciferase expression plasmid, and 250 ng of either an empty myc-pcDNA3.1 vector or a plasmid expressing SARS-CoV-2 PLpro domain of Nsp3 (amino acids 745-1061), using the TransIT-mRNA Transfection Kit (Mirus bio). To stimulate signaling, cell media was replaced with media containing the compound and 20 ng/mL tumor necrosis factor alpha TNF-α (R&D Systems). For both assays, cells were harvested 24 hours post transfection using 1x passive lysis buffer and the dual luciferase assay was performed according to instructions from manufacturer (Promega). Luciferase signals were read using a BioTek Synergy H1 plate reader (Thermo Fisher Scientific).
2.4 Pharmacokinetics Analysis in Mice
Adult C57BL/6 male mice received DDL-701 or DDL-750 in DMSO alone or in combination at 30 mg/Kg (each) by oral gavage. Two hours after dosing, mice were anesthetized and blood was collected following transcardial puncture for the isolation of plasma. Mice were then perfused transcardially with saline and brain tissue removed. Levels of DDL-701 and DDL-750 were determined in brain and plasma by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Briefly, a targeted LC-MS/MS assay was developed for each compound using the multiple reaction monitoring (MRM) acquisition method on a 6460 triple quadrupole mass spectrometer (Agilent Technologies) coupled to a 1290 Infinity HPLC system (Agilent Technologies) with a Phenomenex analytical column (Kinetex 1.7 µm C18 100 Å 100 x 2.1 mm). The HPLC method utilized a mixture of solvent A (99.9/1 Water/Formic Acid) and solvent B (99.9/1 Acetonitrile/Formic Acid) and a gradient was use for the elution of the compounds (min/%B: 0/20, 3/20, 19/99, 20/99, 21/20, 35/20). In this assay, detection of fragmented ions originating from each compound at specific chromatographic retention times were utilized to ensure specificity and accurate quantification in the complex biological samples (DDL-701 (M+H)+: 443.1, fragment ions: 211.0/322.1, retention time: 21.6; DDL-750 (M+H)+: 500.2, fragment ions: 111.0/319.1, retention time: 9.1). Chromatographic peak areas from standards made in drug naïve plasma and brain tissue lysates with increasing amounts of DDL-701 and DDL-750 were used to make a standard curve, and the trendline equation was used to calculate the absolute concentrations of each compound.
2.5 Modeling of Compound Docking to the PLpro Active Site
The crystal structure of PLpro protein (PDB ID: 7CMD) with GRL-0617 ligand [30] was used as a basis for docking studies. The binding site was defined by placing GRL-0617 in the center of the grid box of approximately 20Å dimensions. Flare (Cresset Software, v5.0.0) was used for docking modeling. The protein was prepared for docking utilizing an internal optimization module. Three separate docking runs were performed for each compound, each generating unbiased ~200 poses, and each set of poses was evaluated for the best fit. For the final manual expert evaluation, ~5-10 poses were selected and then further narrowed to 1-2 poses with the best scores. Each final pose was then examined to identify likely interactions with protein residues (hydrogen bonding, π-π or hydrophobic interactions, etc.). Orientations of compounds showed strong structural interaction with Tyr268 that lies outside the active site tunnel and were considered as preferred binding site of these molecules through π-π and hydrogen bonding interaction with Tyr268. Such compound binding interaction could interfere with PLpro substrate binding and proteolytic activity.
3. Results
3.1 Screening of Clinical Compound Library in PLpro Assay Reveals Hit DDL-701
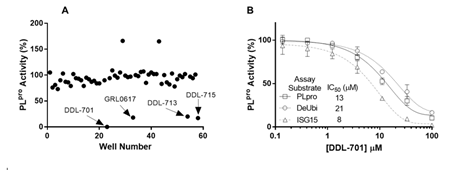
We screened small molecules from a 58-compound custom clinical library that was assembled from commercial sources (see details in Supplementary Table S1) for their ability to inhibit PLpro enzyme activity using a short peptide substrate. As shown in the scatterplot in Figure 1A, the screening revealed 4 hits that inhibit PLpro activity > 80 %: eltrombopag (DDL-701), the known PLpro inhibitor GRL-0617, thrombopoietin receptor agonist-1 (TPO-1 agonist, DDL-713), and zafirlukast (DDL-715) a potent cysteinyl leukotriene receptor antagonist [31]. Interestingly, two compounds – the antipsychotic drug fluspirilene and the leukotriene receptor antagonist montelukast – increased PLpro activity (see compound well # in Supplementary Table S1). To further validate DDL-701 as a PLpro inhibitor, we assessed its dose-response inhibition of PLpro activity, using the PLpro peptide substrate, along with the deubiquitinase and deISGylase substrates (Figure 1B) which showed IC50 values of 13, 21, and 8 μM, respectively. Dose-response curves for PLpro inhibition by the other hits from screening were also generated and displayed weaker potency than DDL-701: DDL-713 (IC50PLpro~54 µM) and DDL-715 (IC50PLpro~24 µM) (Supplementary Figure S1).
3.2 DDL-701 and Nirmatrelvir (DDL-750) Show Sustained Protease Activities in Combination
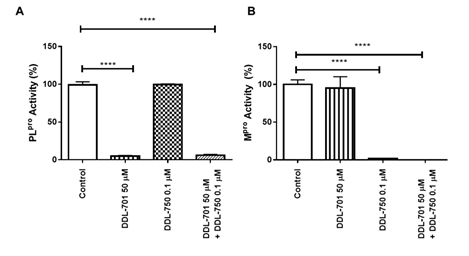
For DDL-701 to be efficacious in protease inhibitor cocktail therapy, it is important that its activity be sustained in the presence of an Mpro inhibitor such as nirmatrelvir (DDL-750). As shown in Figure 2A, inhibition of PLpro by DDL-701 is almost complete at 50 μM and the inhibition is not affected when it is used in combination with DDL-750 in the protease assay. Similarly, DDL-750 Mpro inhibitory activity was sustained in the presence of DDL-701 (Figure 2B). DDL-701 did not show activity in an Mpro activity assay.
Figure 2: DDL-701 and DDL-750 alone and in combination in the PLpro and M pro assays. The (A) PLpro and (B) Mpro assays with DDL-701 and DDL-750 each alone and in combination at the concentrations shown. Statistical analysis performed using one-way ANOVA with post-hoc comparison of each treated well to the control, where ****p < 0.0001.
3.3 DDL-701 Activity Partially Restores Interferon-β (IFN-β) induction in vitro
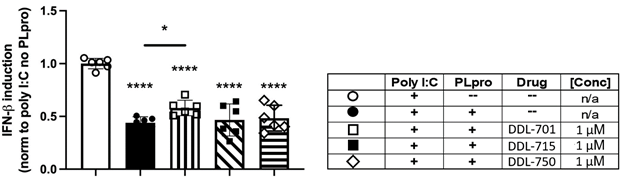
Because PLpro activity is reported to inhibit the host cell IFN-β antiviral response [6], we sought to determine if the PLpro inhibitors can rescue IFN-β induction. HEK-293T cells were co-transfected with an IFN-β luciferase reporter and SARS-CoV-2 Wuhan strain PLpro, and stimulated with polyinosinic:polycytidylic acid (poly I:C) to mimic the double stranded RNA (dsRNA) that positive-sense, single-stranded RNA viruses such as SARS-CoV-2 form during viral replication. As shown in Figure 3, expression of PLpro reduces IFN-β induction by > 50%, and treatment with DDL-701 at 1 µM modestly yet significantly rescued IFN-β induction (p = 0.042). Higher concentrations did not rescue induction. The lack of induction at the higher concentration could be a result of compound interference with the dual transfected HEK cell assay. Although eltrombopag is also known to target other intracellular functions that may contribute to the induction activity seen at 1uM (32), however, most likely the observed induction of IFN-β in the transfected HEK cells is through inhibition of the PLpro activity. Neither DDL-715 nor DDL-750 rescued IFN-β induction at any dose (Supplementary Figure S2A). PLpro activity has also been reported to antagonize NFκB signaling [33], therefore an NFκB assay was also performed, but no rescue of NFκB signaling was observed by any compound tested under the conditions of the assay (Supplementary Methods and Figure S2B).
Figure 3: DDL-701 partially restores interferon-beta (IFN-b induction by poly I:C in the presence of PLpro. Polyinosinic:polycytidylic acid (poly I:C) was used to stimulate IFN-β induction in HEK-293 cells. Transfection with a reporter construct expressing Wuhan strain PLpro (see legend in table) significantly decreases IFN-β induction as compared to the poly I:C only control, which is partially restored by the PLpro inhibitor DDL-701, but not by DDL-715 or Mpro inhibitor DDL-750, all at 1 μM. Statistical analysis performed using one-way ANOVA with Dunnett’s post-hoc comparison of the poly I:C only control to all other groups and Sidak’s post-hoc comparison of poly I:C + PLpro no drug (--) to Poly I:C + PLpro + DDL-701, 715 or 750 where ****p < .0001 and *p ≤ .05.
3.4 DDL-701 is Brain-Permeable in Mice
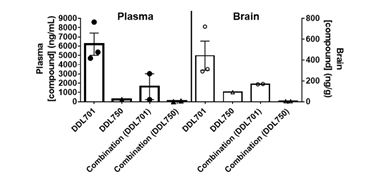
To determine if DDL-701 as part of a protease inhibitor cocktail has the potential to be effective against SARS-CoV-2 infection of the central nervous system (CNS), a pharmacokinetic study was performed in mice. Animals received DDL-701 or DDL-750 alone or in combination both at 30 mg/Kg by oral dosing, and brain and plasma levels assessed. As shown in Figure 4, 2 hours after oral treatment, brain levels of DDL-701 were ~1 μM (~442 ng/g) while DDL-750 (nirmatrelvir) reached a concentration of ~0.18 μM (~91 ng/g). DDL-701 has >5 fold higher brain penetration than nirmatrelvir. In plasma, DDL-701 and DDL-750 levels were ~14 μM (~ 6,214 ng/mL) and ~0.5 μM (~234 ng/mL) respectively. In combination, the levels in plasma and brain were lower for both drugs; DDL-701 levels in plasma were ~4 μM (~1,628 ng/mL) and in brain ~0.4 μM (~ 169 ng/g), and DDL-750 levels were ~0.1 μM (~55 ng/mL) and ~0.1 μM (~ 59 ng/g) in plasma and brain, respectively. The drug level of DDL-701 in the mouse plasma was close to the IC50 for PLpro inhibition, while the levels in the brain were close to the 1 μM dose that showed IFN-β induction. In combination with DDL-750 both plasma and brain levels of DDL-701 in mice were decreased. The pilot pharmacokinetic study was done in a small group of mice and the results could be affected by mouse-to-mouse variability. The mice studies were done under an approved animal study protocol.
3.5 Modeling and Docking Reveal DDL-701 and Known PLpro Inhibitor GRL-0617 Interact with Residues near the Active Site
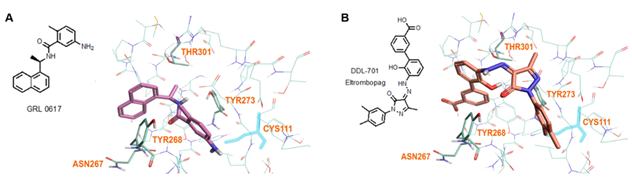
Modeling by docking of DDL-701 with PLpro was compared with that of the known PLpro inhibitor GRL-0617 (PDB ID: 7CMD) [34] using Cresset software. The docking data suggests that like GRL-0617, DDL-701 (Figure 5A and 5B, respectively) does not interact with the active site cysteine-111 residue, but rather can bind effectively to the site around tyrosine-268 which lies just outside the tunnel containing the active site residue Cys111. DDL-715 and the reported PLpro inhibitor losartan [22] bind the enzyme like DDL-701, at the site around Tyr268 and at the entrance of the active site tunnel leading to Cys111 (Supplementary Figures S3 and S5, respectively). Similarly, docking of DDL-713 which is a TPO agonist like DDL-701 (Supplementary Figure S4) binds around Tyr268 located outside the catalytic-site cavity containing the key residue Cys111.
Figure 5: Model of GRL-0617 (PLpro inhibitor) and DDL-701 (Eltrombopag) in the active site of PLpro. (A) X-ray crystallography structure of PLpro and GRL-0617 (PDB ID: 7CMD) is shown. (B) DDL-701 binds similar to GRL-0617, based on the PLpro amino acid residues marked in the vicinity of these two ligand binding sites. Both molecules bind at the entrance of the active site tunnel around Tyr268 and distal from the active site residue Cys111.
4. Discussion
The FDA-approved drug DDL-701, identified in our screening of a library of clinical compounds, shows micromolar PLpro inhibition and elicits partial restoration of IFN-β induction in vitro. As shown in Figure 2, while DDL-701 only inhibits PLpro and not Mpro the combination protease inhibitor cocktail of DDL-701 and nirmatrelvir (DDL-750) effectively inhibits both PLpro and Mpro enzyme activity. DDL-701 has an advantage for use in immediate clinical testing because it is already approved for thrombocytopenia (available in the US under the brand name PROMACTA and outside the US as REVOLADE), and human pharmacokinetic (PK) data is available. These reported PK data indicate that a single oral dose of 75 mg DDL-701 in tablet form results in plasma Cmax levels in the range of 25-36 μM with an AUC of 168 μg-hour/mL [35], a level that is above its IC50 of ~ 13 μM for inhibition of PLpro enzyme. DDL-701 thus would be likely to exert PLpro inhibition in vivo in human patients after an oral dose and could show robust antiviral efficacy most effectively in a protease inhibitor cocktail with a Mpro inhibitor.
Another PLpro inhibitor identified from our screening, DDL-715, is less potent than DDL-701 and in human achieves a Cmax of ~ 0.5 μM in plasma after oral dosing [31]. This is similar to what was shown with another PLpro inhibitor losartan that also has a reported IC50 of ~13 μM for PLpro inhibition [21]. After oral dosing with losartan, however, the Cmax in plasma is only ~ 0.5 μM, well below its IC50 for PLpro inhibition. DDL-715 and Losartan, in our modeling, also can bind the active site region similarly to DDL-701 (Supplementary Fig S5). The PLpro inhibitor tropifexor, a potent Farnesoid X Receptor agonist, is reported to have a PLpro IC50 ~6 μM [25]. The binding of tropifexor in our modeling again is similar to DDL-701 around the Tyr268 site (Supplementary Figure S6). The Cmax in human plasma with an approved oral dose is <0.1 μM, well below its PLpro IC50. All of these data show that DDL-701 has an advantage, in that after a clinically used oral dose of the drug it is possible to achieve plasma levels above the PLpro IC50, making it a promising candidate to repurpose as part of the protease inhibitor cocktail with a Mpro inhibitor such as nirmatrelvir for treatment of COVID-19. Here, we also report that our pilot PK studies in mice show DDL-701 is brain-permeable, which suggests it has potential to counter the effects of SARS-CoV-2 infection and potential complications in the CNS [36]. Nirmatrelvir had lower brain-permeability, but it is highly potent for the Mpro enzyme with an IC50 of ~3 nM [15], therefore it may be effective in brain as part of the protease inhibitor cocktail. The establishment of such efficacy awaits testing in animal models of SARS-CoV-2 infection and/or in clinical trials.
As further evidence of its potential in a protease inhibitor cocktail, DDL-701 has previously been reported to have antiviral efficacy in Vero E6 cells infected with SARS-CoV-2 with an EC50 of 8.2 μM and EC90 of 9.5 μM [20], although a mechanism of action for eltrombopag in that report is not described. In Vero E6 normal cells treated with DDL-701 the CC50>50μM was reported [20]. The reported antiviral efficacy in SARS-CoV-2 infected VeroE6 cells for nirmatrelvir was EC50 of 74 nM while the reported EC90 was 155 nM and in normal cells the CC50 was > 100μM [15]. In a separate study, eltrombopag was reported to show the ability to bind and reduce the stability of the spike protein-ACE2 complex [ 37], which may contribute to its antiviral efficacy. Importantly, the plasma Cmax level of DDL-701 after a single oral dose in humans is well above its EC50 for inhibition of SARS-CoV-2 infection. Thus, the oral dose of 75 mg DDL-701 as a protease inhibitor cocktail with Paxlovid in COVID-19 patients has the potential to produce robust antiviral efficacy. These studies await confirmation through clinical testing.
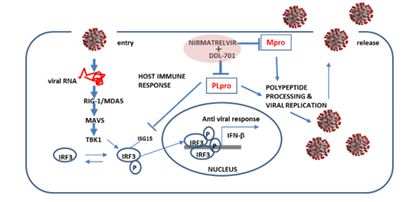
Ideally, the PLpro inhibitor DDL-701 used in a protease inhibitor cocktail with the Mpro nirmatrelvir would result in induction of host antiviral activity, in addition to reducing viral polypeptide processing and replication. During viral infection there is induction of host antiviral response, cytosolic viral RNA forms a complex with the proteins RIG-1/MDA5, interacts with the molecular cascade involving MAVS, TBK1 and IRF3, leading to phosphorylation and nuclear translocation of IRF3 inducing transcription of type I interferons [6], this response can be antagonized by PLpro (Figure 6). By interfering with PLpro cleavage of ISG15 and thus IRF3 phosphorylation [38], DDL-701 may restore IFN-β signaling in vivo, a potential effect supported by our data showing increased IFN-β induction in vitro. The lack of response to NF-KB signaling could be due differential mechanisms by which its signaling is antagonized by PLpro [33, 39]. Confirmation of these potential mechanisms awaits further study.
Figure 6: Putative restoration of host immune responses by a DDL-701/nirmatrelvir cocktail. PLpro alters IRF3 phosphorylation activity and the IFN-β host response. In combination with an Mpro inhibitor such as nirmatrelvir, it could help restore the host cell immune response in addition to reducing viral replication.
While the SARS-CoV-2 virus does not easily enter the brain, it has been reported to damage endothelial cells in the blood brain barrier (BBB) leading to inflammation and brain injury [40]. Knowledge concerning the impact of SARS-CoV-2 infection on the CNS and cerebrovasculature is limited and still being elucidated. The entry of the virus into the brain through the olfactory nerve endings and at the blood-CSF barrier (choroid plexus) is postulated [36, 41]. Evidence also suggests that the virus can infect astrocytes in the CNS and could lead to some of the observed neurological symptoms such as fatigue, depression and brain fog [40]. Our pilot pharmacokinetic studies in mice herein suggest DDL-701 is brain-permeable, and while the brain permeability of nirmatrelvir is low, given its nanomolar potency in inhibiting viral replication [15] as part of the protease inhibitor cocktail it may have the potential to lower the risk of CNS complications.
5. Conclusions
Based on our findings, DDL-701 is a PLpro inhibitor with sustained inhibitory activity in the presence of Mpro inhibitor nirmatrelvir, it has previously been reported to achieve plasma levels that are likely to elicit both PLpro inhibition and antiviral efficacy. It is a promising candidate for further clinical study in combination with a Mpro inhibitor as a protease inhibitor cocktail for the treatment of SARS-CoV-2 infection. DDL-701’s brain permeability suggested by our studies further shows its potential to reduce CNS symptoms of the infection. DDL-701 has the advantage of being an FDA approved drug for thrombocytopenia and can be readily repurposed using the already available oral tablet that could be reformulated in combination with oral Paxlovid in the current five-day treatment regimen. Such a protease inhibitor cocktail presents the possibility of a more effective treatment for COVID-19 patients worldwide that has the potential to reduce hospitalization, deaths resulting from the disease, speed up post-infection recovery, reduce the risk of rebound, reduce antiviral resistance and CNS complications.
Patents
There is a patent application filed from the work reported in this manuscript - UCLA Case No. 2022-177.
Supplementary Materials
The following supporting information can be downloaded at: https://www.fortunejournals.com/supply/ACBR_7249.pdf Table S1; Figure S1-S6.
Author Contributions
Substantial contribution to conceptualization and design, J.C., M.L., R.D. and V.J.; substantial contribution to methodology, acquisition, analysis and interpretation of data, J.C., B.J., P.A., K.M.E, W.C., P.F., V.A., and C.Y..; resources, R.D., M.L., A.J.O., and V.J.; drafting the work and revising it critically for important intellectual content, J.C., P.A., B.J., R.D., M.L., A.J.O., W.C., and V.J.; supervision, M.L., R.D., and V.J.; project administration, M.L., R.D., and V.J.; funding acquisition, M.L., R.D., and V.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by pilot grants from the David Geffen School of Medicine at UCLA; pilot grant (OCRC #20-69) to V.J. and the pilot grant (OCRC #20-16) to R.D, that enabled the screening and identification of the drug candidates. The APC was funded by an unrestricted fund from the John lab.
Acknowledgments
We would like to thank the David Geffen School of Medicine at UCLA for the pilot grants to V.J. and R.D. supporting this research. M.L. was supported in part by the Johanna and Joseph H. Shaper Family Chair.
Conflicts of Interest
The authors declare no conflict-of-interest disclosures.
References
- Soneji S. Population-level mortality burden from novel coronavirus (COVID-19) in Europe and North America. Genus 77 (2021): 7.
- Li X. Omicron: Call for updated vaccines. J. Med. Virol 94(2022): 1261-1263.
- Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res 149 (2018): 58-74.
- Ratia K, Kilianski A, Baez-Santos YM, et al. Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating Activity of SARS-CoV Papain-Like Protease. PLoS Pathog 10 (2014): e1004113.
- Yadav R, Chaudhary JK, Jain N, et al. Role of Structural and Non-Structural proteins and therapeutic targets and SARS-CoV-2 for COVID-19. Cell 10 (2021): 821.
- Shin D, Mukherjee R, Grewe D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587 (2020): 657-662.
- Matthews K, Schäfer A, Pham A, et al. The SARS coronavirus papain-like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol. J 11 (2014): 209.
- Freitas B, Durie IA, Murray J, et al. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis 8 (2020): 2099-2109.
- Li C, Debing Y, Jankevicius G, et al. Viral Macro Domains Reverse Protein ADP-Ribosylation. J. Virol 90 (2016): 8478-8486.
- Malgras M, Garcia M, Jousselin C, et al. The Antiviral Activities of Poly-ADP-Ribose Polymerases. Viruses 13 (2021): 582.
- Sun L, Xing Y, Chen X, et al. Coronavirus Papain-like Proteases Negatively Regulate Antiviral Innate Immune Response through Disruption of STING-Mediated Signaling. PLoS ONE 7 (2012): e30802.
- Mielech AM, Kilianski A, Baez-Santos YM, et al. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450–451 (2014): 64-70.
- Ma-Lauer Y, Carbajo-Lozoya J, Hein MY, et al. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PL pro via E3 ubiquitin ligase RCHY1. Proc Natl Acad Sci U S A 113 (2016): E5192-201.
- Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369 (2020): 718-724.
- Owen DR, Allerton CMN, Anderson AS, et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 374 (2021): 1586-1593.
- Gupta K, Strymish J, Stack G, et al. Rapid relapse of symptomatic omicron SARS CoV-2 infection following early suppression with nirmatrelvir/ritonavir. 26 April 2022, PREPRINT (Version 1) available at Research Square (2022).
- Carlin AF, Clark AE, Chaillon A, et al. Virologic and Immunologic Characterization of COVID-19 Recrudescence after Nirmatrelvir/Ritonavir Treatment. 18 May 2022, PREPRINT (Version 1) available at Research Square (2022).
- Sacco MD, Hu Y, Gongora MV, et al. The P132H mutation in main protease of omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition. Cell Res 32 (2021): 498-500.
- Zhou Y, Gammeltoft KA, Ryberg LA, et al. Nirmatrelvir Resistant SARS-CoV-2 Variants with High Fitness In vitro. BioRxiv (2022).
- Jeon S, Ko M, Lee J, et al. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob Agents Chemother 64 (2020): e00819-20.
- Nejat R, Sadr AS, Freitas B, et al. Losartan inhibits SARS-CoV-2 Replication in vitro. J. Pharm Pharm Sci 24 (2021): 390-399.
- Jamir E, Sarma H, Priyadarsinee L, et al. Applying polypharmacology approach for drug repurposing for SARS-CoV-2. J. Chem. Sci 134 (2022): 57.
- Lewis DSM, Ho J, Wills S, et al. Aloin isoforms (A and B) selectively inhibits proteolytic and deubiquinating activity of PLpro of SARS-CoV-2 in vitro. Sci. Rep 12 (2022): 2145.
- Napolitano V, Dabrowska A, Schorpp K, et al. Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses. Cell Chem Bio 29 (2011): 774-784.
- Chunlong M, Hu Y, Wang Y, et al. Drug-Repurposing Screening Identified Tropifexor as a SARS-CoV-2 Papain-like Protease Inhibitor. ACS Infectious Disease 8 (2022): 1022.
- Narayanan A, Narwal M, Majowicz SA, et. Al., Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in cell protease assay. Commun. Biol 5 (2022): 169.
- Kulandaisamy R, Kus, hwaha T, Dalal Aet al. Repurposing of FDA approved Drugs against SARS-CoV-2 papain-like protease: Computational, Biochemical and in vitro Front. Microbiol 13 (2022): 877813.
- Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science 368 (2020): 409-412.
- Mellott DM, Tseng C, Drelich A, et al. A clinical-stage cysteine protease inhibitor blocks SARS-CoV-2 infection of Human and Monkey Cells. ACS Chem. Biol 16 (2021): 642-650.
- Fu Z, Huang B, Tang J, et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat Commun 12 (2021): 488.
- Murata Y, Sugimoto O. Zafirlukast (Accolate): a review of its pharmacological and clinical profile. Nihon Yakurigaku Zasshi 119 (2002): 247-258.
- Guan Y, Hasipek M, Jiang D, et al. Eltrombopag inhibits TET dioxygenase to contribute to hematopoietic stem cell expansion in aplastic anemia. J. Clin Invest 132 (2022): e149856.
- Frieman M, Ratia K, Johnston RE, et al. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol 83 (2009): 6689-6705.
- Gao X, Qin B, Chen P, et al. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm Sin B 11 (2021): 237-245.
- Matthys G, Park JW, McGuire S, et al. Clinical pharmacokinetics, platelet response, and safety of eltrombopag at supratherapeutic doses of up to 200 mg once daily in healthy volunteers. J Clin Pharmacol 51 (2011): 301-308.
- Gupta M, Weaver D. COVID-19 as a trigger of Brain Autoimmunity. ACS Chem Neurosci 12 (2021): 2558.
- Feng S, Luan X, Wang Y, et al. Eltrombopag is a potential target for drug intervention in SARA-CoV-2 spike protein. Infect Genet Evol 85 (2020): 104419.
- GuanQun L, Lee J, Parker ZM, et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol 6 (2021): 467-468.
- Brisse M, Ly H, et al. Comparative Structure and function analysis of the RIG-1-like receptors: RIG-1 and MDA5. Front. Immunol 10 (2019): 1586.
- Boldrini M, Canoll PD, Klein RS, et al. How COVID-19 affects the brain. JAMA Psychiatry 78 (2021): 682-683.
- McQuaid C, Brady M, Deane R, et al. SARS-Cov-2: is there neuroinvasion? Fluids Barriers CNS 18 (2021): 32.