Human Lung Local Immunity to Influenza Virus-Role of Lung Antibody and Resident Memory T Cells
Article Information
Renee WY Chan1,2,3,4*, Sophie A Valkenburg5,6 , Joanne HM Fong1,2, Man Chun Cheung5, Denise IT Kuok5, Md A Islam3, JSM Peiris5,6, John M Nicholls7, LLM Poon5, Michael CW Chan5
1Department of Paediatrics, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong
2CUHK-UMCU Joint Research Laboratory of Respiratory virus and Immunobiology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong
3Hong Kong Hub of Paediatric Excellence, The Chinese University of Hong Kong, Hong Kong
4Laboratory for Paediatric Respiratory Research, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong
5Division of Public Health Laboratory Sciences, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong
6HKU Pasteur Research Pole, The University of Hong Kong, Hong Kong
7Department of Pathology, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong
*Corresponding author: Renee WY Chan, Department of Paediatrics, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong.
Received: September 13, 2023; Accepted: September 19, 2023; Published: January 20, 2024
Citation: Renee WY Chan, Sophie A Valkenburg, Joanne HM Fong, Man Chun Cheung, Denise IT Kuok, Md A Islam, JSM Peiris, John M Nicholls, LLM Poon, Michael CW Chan. Human Lung Local Immunity to Influenza Virus-Role of Lung Antibody and Resident Memory T Cells. Archives of Clinical and Biomedical Research. 8 (2024): 01-09.
Share at FacebookAbstract
Human lung explant cultures have been used extensively to risk assess the replication competence and disease severity when a new respiratory virus emerges. In the 2009 H1N1 pandemic, we observed an interesting pattern of a steep increase in H1N1pdm virus replication in the human lung ex vivo culture collected from April to July 2009 to the almost flattened replication curve in the period of August 2011 to July 2012 onwards. As the lung explant system is isolated from the circulatory immunity, we aimed to determine the effect of the lung local immunity towards the influenza virus by measuring the antibody levels in the lung epithelial lining fluid (ELF) and the involvement of tissue-resident memory T cells. Lung ELF contained IgA and measurable IgG however, it is not sufficient to neutralize the influenza virus used in the current study. CD8+ T resident memory (TRM) cell and the CD4+TRM cell can be found in the lymphocyte population isolated from the lung. Interestingly, TRM CD8+Tet+ was associated with a trend of reduced replication competence of H1N1pdm in the lung explant culture but not in seasonal H1N1. The presence of influenza-strain-specific CD8+TRM might reduce the virus replication competence in the local epithelium.
Keywords
Human lung explant culture; Lung local immunity; CD8+ resident memory T cells
Article Details
Introduction
Influenza viruses belong to the Orthomyxoviridae family and are characterized by a segmented, negative sense, and single-stranded RNA (ssRNA) genome. Influenza virus disease has caused significant public health concerns, and hastened pandemics several times over the past 100 years, specifically in 1918, 1957, 1968 and 2009 [1]. Although the seasonal influenza virus usually not lead to fatality, its high transmissibility in humans makes the disease burden substantial. Influenza virus is not yet eradicable because of its vast reservoir in animals and high mutation rates, imposing the threat of the re-emergence of old pandemic viruses and the emergence of novel viruses with pandemic potential [2]. Various study models have been used, including cell cultures and animal models, to study the tissue tropism and pathogenesis of these viruses. Amongst all the available studying models, primary human respiratory organ culture is regarded as one of the Influenza Risk Assessment Tool (IRAT) [3] and takes an important role in providing clinically relevant data to reflect host responses of humans upon influenza infection. We have previously established and characterized the human lung explant culture model since 2006 and successfully used this lung explant culture model to address important questions regarding the tissue tropism of highly pathogenic avian influenza H5N1 and H7N9 viruses [4,5], the 2009 pandemic H1N1 virus [6], the MERS-CoV [7], influenza B virus [8] and SARS- CoV-2 [9] and also employed as a risk assessment model to evaluate the pandemicity for the swine [10] and avian viruses that isolated from Hong Kong surveillance program to start a new pandemic in humans. This ex vivo infection series is the largest and most comprehensive study to date. The tropism and replication kinetics data correlate well with the ferret transmission study [11]. In our human lung explant model, the biological tissue under examination was the lung tissue which has been isolated from the systemic circulation. Therefore, we first hypothesized that the Immunoglobulin (Ig) in the alveolar Epithelial Lining Fluid (ELF) could be one of the contributing factors to the change in the replication kinetics. However, the Ig concentration in alveolar ELF is poorly understood. The best estimate is based on the calculation of IgG level in bronchoalveolar lavage (BAL) sample which also takes into account the ratio between serum and BAL urea levels [12]. Moreover, there has not been a comprehensive study regarding the quantity of Ig and also influenza-specific neutralizing antibody in alveolar space, which is of relevance to the current explant culture model. In addition, as the majority of circulating immune cells and lymph nodes are not present in this ex vivo setup, we also investigated if a set of specific cellular components yields this viral strain-specific immunity over time. Lung resident immune cells with adaptive immune responses are highly relevant [13]. Furthermore, a third population of memory T cells named resident memory T (TRM) cells, have been identified, especially in secondary lymphoid organs and barrier tissues [14,15]. They usually do not recirculate in the blood and persist permanently in these tissues and can stand for a rapid immune response to restrict infection within the local tissue sites [16,17]. In this study, we examined the presence of resident immune cells, including lung-specific resident memory cells and their association with viral strains’ specific immunity in explant lung culture.
Materials and Methods
Viruses
Pandemic virus A/HongKong/415742/09 (H1N1pdm) and seasonal viruses A/HongKong/54/98 (H1N1) were used. The viruses were initially isolated and passaged in Madin-Darby canine kidney (MDCK) cells. The virus stock was aliquoted and then titrated to determine a tissue culture infection dose of 50% (TCID50) in MDCK cells.
Thin slice human lung explant cultures
Fresh lung tissues were obtained from patients undergoing elective surgery in the Queen Mary Hospital in Hong Kong and were removed as part of clinical care but surplus for routine diagnostic requirements as detailed previously [8].
Briefly, the tumour-free regions of the residual lung tissues were transported in a sterile bottle to our laboratory within 1h. The lung was perfused with cold transport medium (sterile PBS (pH 7.2) containing 200 μg/ml of gentamicin, 100 U/ ml of penicillin, 100 μg/ml of streptomycin, and 2.5 μg/ml of amphotericin B) using clear tracheal tube 2.0mm (Ruschelit) through bronchiole until the lobe was fully expanded. The cold transport medium was then aspirated and replaced by 1% agarose (Sigma, A0701) in PBS (Sigma). This tissue was cut into cubes and further embedded in 4% agarose. The agarose- embedded lung tissue was cut in slices, using a cryotome blade with a thickness <1mm. The thin slices of the lung were cut using a disposable biopsy punch (Miltex) in a diameter of 5mm and placed on a sterile surgical sponge (Simport) inside a 12-well plate. 1.5 ml of the corresponding medium was added into each well with the surgical sponge float with the explants on the medium to create an air-liquid interface. The medium was changed every hour in the first four hours and incubated at 37°C in a water-jacketed incubator with a 5% CO2 supply to allow the melting and removal of agarose within the lung slice. Lung slide culture medium (LSM) composition: Minimal essential medium (Sigma, St. Louis, MO) supplemented with 1.0 μg of bovine insulin/ml (Sigma), 0.1 μg/ml of hydrocortisone (Sigma), 0.1 μg/ml of vitamin A (Sigma), 200 μg/ml of gentamicin, 100 U/ml of penicillin, 100 μg/ml of streptomycin and 1.25 μg/ml of amphotericin B, as described [18]. Thin lung slice infection was conducted as previously described [6]. The use of human biospecimens had been previously approved by the local institutional review board (UW 13-064).
Alveolar epithelial lining fluid (ELF) extraction and immunoglobulin measurements
Once the fresh lung tissue arrived at our laboratory, 3g was used for the extraction of alveolar ELF. This portion was cut into thin slices (< 1mm) using microtome with minimal mechanical damage to the tissue. The slices were put into 3 ml of cold PBS together with a magnetic stirrer and the solution was stirred on ice for 30 min. The supernatant of this tissue solution was recovered by centrifugation at 1100 rpm for 5 min would be the alveolar ELF. The supernatant was aliquoted into a small volume and stored at -20°C. IgA and IgG levels of the lung wash and lung homogenate were detected by human IgA-ELISA Kit (Cygnus, F165, detection range 0.5 – 50 ng/ml) and human IgG-total ELISA Kit (Cygnus, F160, detection range 0.5 – 50 ng/ml) in duplicate, respectively.
Haemagglutinin inhibition (HI) assay and Microneutralization (MN) test
The alveolar ELF and serum sample of the same patient who provided the lung tissue was collected for HI and MN tests towards the 2009 pandemic virus and seasonal influenza virus as described [19].
Statistical analysis
The virus replication yield of H1N1 and H1N1pdm was expressed as the area under curve (AUC) below the virus replication kinetic curve from 24-to-48 hours post-infection as described [7]. Paired t-test was performed to compare the viral yield between H1N1 and H1N1pdm. The association between AUC and HI and MN titers of serum was evaluated by linear regression. The association between AUC and the percentage of Tet+CD8+ cells, Tet mean fluorescent intensity and TRM CD8+ Tet+ cells were tested by linear regression. Differences were deemed to be significant if p-value < 0.05. The analyses were performed with GraphPad Prism version 8.4.3 for Mac.
Results
Virus replication kinetics
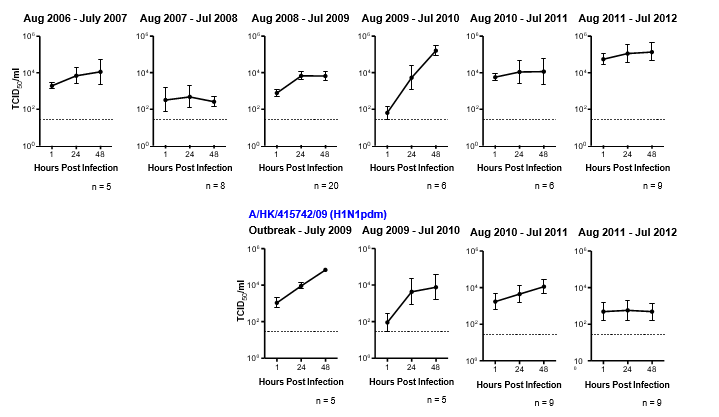
We found that not every influenza virus gives a stable pattern of replication kinetics every year. We observed a productive viral yield of an H1N1pdm virus in the initial phase of the outbreak, i.e. April – July 2009 followed by a drastic decrease in the replication competence in the period of August 2011 to July 2012 (Figure 1). We speculated this phenomenon is mainly contributed by the host immunity, which may relate to their exposure to the virus or vaccination.
IgA and IgG concentrations of alveolar ELF
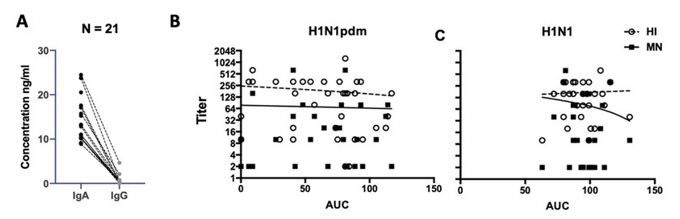
IgA and IgG were measured in 21 alveolar ELFs. The total IgA concentrations were significantly higher than the total IgG concentration in alveolar ELF. IgG concentrations in the alveolar ELF were mostly below 20 ng/ml (Figure 2A).
HI and MN titer against seasonal H1N1 and H1N1pdm viruses of alveolar ELF and serum
We performed the HI assay to determine the concentration of the influenza A virus-specific antibody against A/HK/54/98 (H1N1) and A/HK/415742/09 (H1N1pdm). However, from the 21 lung wash samples tested, only two of them have a HI titer of 1:20 to seasonal H1N1 (A/HK/54/98) and one of them has a HI titer of 1:10 to H1N1pdm (A/HK/415742/09), others were all negative in the HI test of these three strains of influenza viruses. With the limited data points, we did not test for the association between the virus replication yield and the HI titer of the alveolar ELF. Therefore, it suggested that the local immunity of explant culture might not contribute to the Ig present in the alveolar ELF.
Figure 1: Data of human lung explant cultures infected with 106 TCID 50/ml of seasonal H1N1 virus (A/HongKong/54/1998) and pandemic H1N1 virus (A/HK/415742/2009) from 2006 to 2012. Virus replication kinetics of these viruses in the human lung was determined by virus titration at 1, 24, and 28 hours post-infection and expressed in TCID50/ml. The chart showed the pooled data from different patients denoted with n number shown below each graph for clarity. The dotted line indicates the detection limit of the viral titration assay.
Figure 2: (A) IgA and IgG levels of the lung ELF (n=21) were detected by human IgA-ELISA Kit (Cygnus, F165, detection range 0.5 – 50 ng/ml) and human IgG-total ELISA Kit (Cygnus, F160, detection range 0.5 – 50 ng/ml) in duplicate, respectively. The dotted line connected the result of IgA and IgG of the same sample. Wilcoxon matched-pairs signed rank test identified significantly higher IgA levels than IgG of the same lung ELF was identified (p < 0.0001) (B) H1N1pdm and (C) H1N1 infectious viral load from 24 to 72 hpi were expressed in Area Under Curve value (AUC) and plotted against the corresponding serum HI (open circle) and MN titers (square). Linear regression showed no statistical association between the HI titer and the MN titer to the virus replication competence in the ex vivo lung cultures.
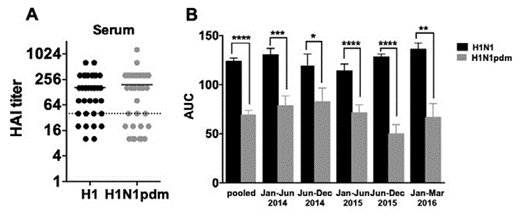
To assess if the circulating immunity has a role in the local immunity in the lung explant setup, the serum sample of the same patient was collected for the HI test towards seasonal H1N1 and 2009 H1N1 pandemic virus. The serum titer showed no difference between H1N1pdm and H1N1 seasonal, with a mean value of 1:190 and 1:164, respectively (n = 37). If we used the protective titer of 1:40 as the cutoff value for a 50% reduction in the prevalence of infection [20], all of the samples collected after 2015 exceeded this threshold (Figure S1A). However, it is worth noting that this 1:40 cutoff value is controversial as it varies across age groups. Moreover, an age-adjusted value cutoff value for H1N1 would be much higher than 1:40 was defined in 1970 for 50% protection [21]. The overall and time-stratified replication efficiency of the seasonal H1N1 virus was higher than H1N1pdm (Figure S1B, p < 0.0001, pair-t test, two-tailed).
The serum HI and MN titer showed no difference between H1N1pdm and seasonal H1N1, with a mean HI titer of 1:190 and 1:164, respectively (n = 37) and a mean MN titer of 1:65 and 1:76, respectively (n = 35). With the comparable HI titers between seasonal H1N1 and H1N1pdm but a higher viral load of seasonal H1N1 than H1N1pdm in the explant lung infection experiment (Figure S1B), there were no associations identified between the HI and MN titer in the serum to the viral yield in the H1N1pdm (Figure 2B) and seasonal H1N1 (Figure 2C) infected lung explant culture.
A trend of reduced replication competence of H1N1pdm in the lung explant culture with the increased influenza-specific CD8+ T cells
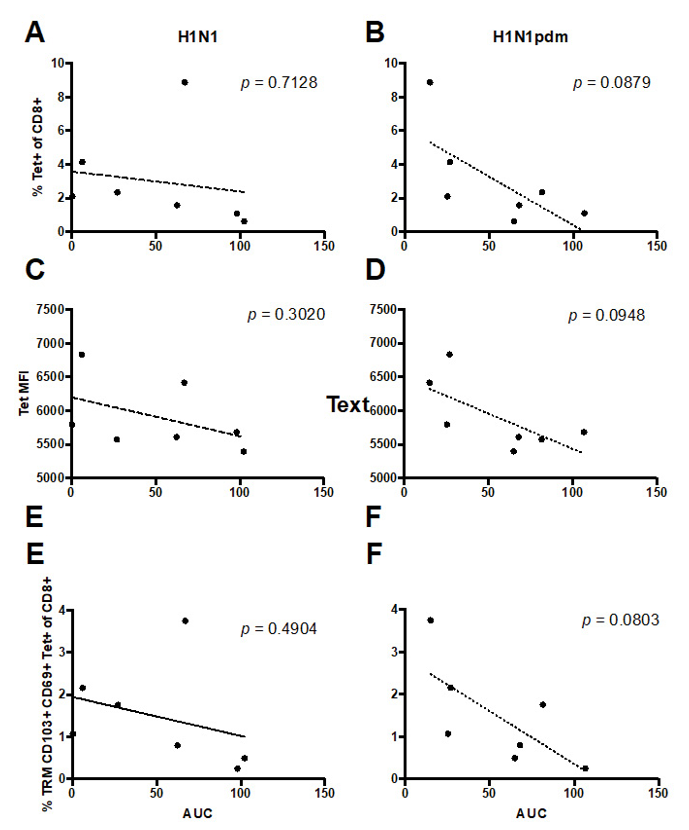
To assess influenza specific T cell resident memory in the lung tissue we used two approaches, intracellular cytokine staining (ICS), which can simultaneously assess influenza specific CD4+ and CD8+ T cell responses by cytokine secretion, and tetramer pools for influenza specific CD8+ T cell responses (Table S1). However, due to technical issues of low cellular viability following ICS of frozen lung samples and a lack of MHCII tetramers for CD4+ T cells, we were unable to obtain data on CD4+ T cell responses. The tetramer pool for CD8+ T cell responses contained 12 tetramers for immunodominant epitopes for HLA supertypes, with an expected coverage of 80% of the population [22]. Lungs were stained for T cell resident memory, defined by the expression of CD69 and CD103 [23], and 3 of 8 donors were HLA-A2+. All subjects had influenza-specific tetramer+ CD8+ T cells, of which 20-77% were also T cell resident memory phenotype, which was also consistent with HLA-A2+ donors. Due to the cross-reactive nature of T cells and the conserved epitopes within seasonal and pandemic H1N1 viruses, we were not able to discern the strain-specific CD8+ T cell responses in our limited sample size. However, there was a trend for the level of replication of pandemic but not seasonal viruses in lung explant cultures to inversely correlate with the magnitude of tetramer-specific and TRM CD8+ T cells (Figure 3). We found that the higher percentage of CD8+ Tet+ (Figure 3B, p = 0.09) and the TRM CD8+ Tet+ (Figure 3F, p = 0.08) had a trend to reduce the replication competence of H1N1pdm in the lung explant culture. In the ICS assay, we stimulated the lung cell suspension at an MOI of 4 and detected the IFNγ+CD4+TRM and the IFNγ+CD8TRM cells at 12 hpi. However, no correlation was identified with the virus replication competence in the lung explant culture.
Discussion
A time-series observation of the dynamic changes of influenza virus replication competence in human lung explant culture initiated this study. The dampened replication
Figure 3: The percentages of (A and B) Tet+ CD8+ cells, (C and D) Tet mean fluorescent intensity (MFI, which positively reflects the level of T cell receptor expression) and (E and F) TRM CD8+ Tet+ cells of the individual donor were plotted against the corresponding (A, C and E) H1N1 and (B, D and F) H1Npdm viral replication competence in the ex vivo lung cultures expressed in AUC from 24 to 48 hours post- infection. Linear regression was performed with the p-values shown in the corresponding figures.
competence of pandemic H1N1 was not likely due to the contribution by the change in innate immune defence mechanisms and assumed that influenza virus receptors, physical and chemical barriers (e.g. surfactant secretion, antiviral pathway) in the lung did not change significantly across these six years (from 2006 to 2012) in the human population, as the mentioned effect would similarly affect the seasonal A/HK/54/1998 (H1N1). Therefore, the involvement of normal structural cells of the lung, including epithelial cells (alveolar type I cells, alveolar type II cells, clara cells, bronchiolar epithelial cells), endothelial cells (lymphatic endothelial, vascular endothelial), alveolar macrophages, fibroblasts and mesenchymal cells were excluded in this study. With the discrete lung explant culture model, which is disconnected from the circulating immunity, we studied if humoral mediators in alveolar ELF and resident memory T cells contribute to the differential replication competence in seasonal and pandemic H1N1 viruses.
In previous studies, the protection of the lung towards influenza virus infection was mainly contributed by IgG rather than IgA [24,25] and confirmed by a meta-analysis study in identified genes responsible for triggering distinct immune responses to inactivated and live-attenuated influenza vaccines [26]. However, in the current setting with thin tissue sectioning, we consistently detected a higher concentration of total IgA than IgG in the alveolar ELF. It could be because IgG antibody response is not long-lasting, with a half-life of weeks, and declines with age, by which most of the samples were obtained from subjects above 60 years old. Moreover, only a limited number of alveolar ELF samples provided a measurable HI titer. The use of PBS to elute alveolar ELF might involve a significant dilution of samples and interfere with the detection accuracy. Absorptive matrices might be considered as an alternative mucosal fluid collection method in future studies [27].
Lung resident immune cells with adaptive immune responses are exceptionally relevant in this context. TRM CD8 T cells persist within the lung [28] and represent a non- circulating population of memory CD8 T cells [29]. Whilst CD4+ T cells may not lodge in the lung but remain in the nearby draining lymph node, we were unable to address CD4+ TRM in our study. Virus-specific CD8+ TRM is located within pulmonary tissue including bronchioles and alveoli, and resident memory T cells are abundant in the human lung [13]. We identified the CD4+ and CD8+ TRM in our lung explant culture. Specific TRM towards seasonal and pandemic H1N1 was further illustrated by the tetramer staining, and appeared to correlate with virus replication in a strain-specific manner and with time from the pandemic in 2009. Whilst our study is limited by a number of factors, it provides an important insight into the specific role of acquired TRM in potentially limiting local virus replication in the lung.
Conclusions
The change in the susceptibility of H1N1pdm virus in the lung explant culture had a weak link with the circulating antibody levels. With the nature of the epithelial lung fluid we obtained from the washing steps, we could not exclude the contamination of cell population from the residual blood while bronchioalveolar lavage would be a better biospecimen to evaluate the local immunity which was not possible in the setting of our study. The H1N1pdm-specific TRM CD8+ activity identified in this study might be due to the prior exposure of the subject to pandemic H1N1 viruses through natural infection. The correlation of CD8 T cell magnitude and extent of virus replication is on the verge of significance, and it would be worth doing more samples for the tetramer staining and replication.
Author Contributions
Conceptualization, R.W.Y.C.; methodology, R.W.Y.C., S.A.V.; formal analysis, R.W.Y.C., S.A.V.; investigation, S.A.V., J.H.M.F., M.C.C., D.I.T.K.; subject recruitment, J.M.N.; writing original draft preparation, R.W.Y.C., S.A.V.,
M.A.I. ; writing, review and editing, S.A.V., J.S.M.P., J.M.N., L.L.N.P, M.C.W.C; supervision, S.A.V., M.C.W.C.; project administration, R.W.Y.C.; funding acquisition, R.W.Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the General Research Fund (Ref. No. 762213) of the Research Grants Council, University Grants Committee, Hong Kong Special Administrative Region.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the HKU/HA HKW Institutional Review Board (UW 13-064).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated are available from the corresponding author upon reasonable request.
Acknowledgements
We thank Prof Maria P Wong, Department of Pathology, The University of Hong Kong for retrieving the serum samples of the corresponding lung tissue donors and Ms Cassia WM Lin, School of Public Health, The University of Hong Kong the biospecimen transportation and the administration of the clinical research ethics application and regular progress reports. We thank Katherine Kedzierksa and Oahn Nyguen, University of Melbourne, for the provision of HLA-I monomers for TRM straining. We thank all the subjects involved in this study for their generosity and support in our research.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. Part of the results was submitted as an abstract to the seventh ESWI Influenza conference ESWI2020.
References
- Centers for Disease Control and Prevention. Pandemic influenza-past pandemics (2022)
- Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic Cell Host Microbe 7 (2010): 440-451.
- Trock SC, Burke SA, Cox Development of Framework for Assessing Influenza Virus Pandemic Risk. Emerg Infect Dis 21 (2015): 1372-1378.
- Nicholls JM, Chan MC, Chan WY, et Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med 13 (2007): 147-149.
- Chan MC, Chan RW, Chan LL, et al. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory Lancet Respir Med 1 (2013): 534-542.
- Chan MC, Chan RW, Yu WC, et al. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory Am J Pathol 176 (2010): 1828-1840.
- Chan RW, Hemida MG, Kayali G, et Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir Med 2 (2014): 813-822.
- Bui CHT, Chan RWY, Ng MMT, et Tropism of influenza B viruses in human respiratory tract explants and airway organoids. European Respiratory Journal 54 (2019): 1900008.
- Hui KPY, Cheung MC, Perera R, et Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med 8 (2020): 687-695.
- Chan RW, Kang SS, Yen HL, et Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva. J Virol 85 (2011): 11581-11587.
- Yen HL, Liang CH, Wu CY, et Hemagglutinin- neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci USA 108 (2011): 14264- 14269.
- Rennard SI, Basset G, Lecossier D, et al. Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of J Appl Physiol 60 (1985): 532-538.
- Purwar R, Campbell J, Murphy G, et Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One 6 (2011): e16245.
- Gebhardt T, Whitney PG, Zaid A, et Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature 477 (2011): 216-219.
- Schenkel JM, Fraser KA, Masopust Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J Immunol 192 (2014): 2961-2964.
- Jiang X, Clark RA, Liu L, et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin Nature 483 (2012): 227-231.
- Wilk MM, Misiak A, McManus RM, et al. Lung CD4 Tissue-Resident Memory T Cells Mediate Adaptive Immunity Induced by Previous Infection of Mice with Bordetella pertussis. J Immunol 199 (2017): 233-243.
- Chakrabarty K, Wu W, Booth JL, et Human lung innate immune response to Bacillus anthracis spore infection. Infect Immun 75 (2007): 3729-3738.
- Tse M, Kim M, Chan CH, et al. Evaluation of Three Commercially Available Influenza A Type-Specific Blocking Enzyme-Linked Immunosorbent Assays for Seroepidemiological Studies of Influenza A Virus Infection in Pigs. Clinical and Vaccine Immunology 19 (2012): 334-337.
- Grund S, Adams O, Wahlisch S, et Comparison of hemagglutination inhibition assay, an ELISA- based micro-neutralization assay and colorimetric microneutralization assay to detect antibody responses to vaccination against influenza A H1N1 2009 virus. J Virol Methods 171 (2011): 369-373.
- Verschoor CP, Singh P, Russell ML, et Microneutralization assay titres correlate with protection against seasonal influenza H1N1 and H3N2 in children. PLoS One 10 (2015): e0131531.
- Grant EJ, Josephs TM, Loh L, et Broad CD8+ T cell cross-recognition of distinct influenza A strains in humans. Nature Communications 9 (2018): 5427.
- Kumar BV, Ma W, Miron M, et Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep 20 (2017): 2921-2934.
- Ito R, Ozaki YA, Yoshikawa T, et al. Roles of anti- hemagglutinin IgA and IgG antibodies in different sites of the respiratory tract of vaccinated mice in preventing lethal influenza pneumonia. Vaccine 21 (2003): 2362-
- Sridhar S, Brokstad KA, Cox RJ. Influenza Vaccination Strategies: Comparing Inactivated and Live Attenuated Influenza Vaccines (Basel) 3 (2015): 373-389.
- Wen F, Guo J, Huang A meta-analysis identified genes responsible for distinct immune responses to trivalent inactivated and live attenuated influenza vaccines. J Cell Physiol 234 (2019): 5196-5202.
- Rebuli ME, Speen AM, Clapp PW, et Novel applications for a noninvasive sampling method of the nasal mucosa. Am J Physiol Lung Cell Mol Physiol 312 (2017): L288-L296.
- Gebhardt T, Wakim LM, Eidsmo L, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex Nat Immunol 10 (2009): 524-530.
- Turner DL, Bickham KL, Thome JJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T Mucosal Immunol 7 (2014): 501-510.
Figure S1: (A) HI titer against H1N1 seasonal influenza virus (black) and H1N1pdm virus (grey). Dotted line denotes the protective titer of influenza found in serum for a 50% reduction in the prevalence of infection [20]. (B) Influenza virus replication competence plotted using the area under curve (AUC) as the overall output. The biological sample collection dates were stratified into six-months durations. Key: *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.0001
Table S1: Selected MHC-I tetramers for CD8+ T cell staining
|
HLA |
Epitope |
Peptide |
|
HLA-A01:01 |
NP44 S7N |
CTELKLNDY |
|
HLA-A01:01 |
NP44 WT |
CTELKLSDY |
|
HLA-A02:01 |
M158 |
GILGFVFTL |
|
HLA-A03:01 |
NP265 |
ILRGSVAHK |
|
HLA-B07:02 |
NP418 |
LPFERATVM |
|
HLA-B07:02 |
NP418 |
LPFDKTTIM |
|
HLA-B35:01 |
NP418 |
LPFERATVM |
|
HLA-B35:01 |
NP418 |
LPFDKTTIM |
|
HLA-B08:01 |
NP225 |
ILKGKFQTA |
|
HLA-B18:01 |
NP219 |
YERMCNIL |
|
HLA-B27:05 |
NP383 |
SRYWAIRTR |
|
HLA-B57:01 |
NP199 |
RGINDRNFW |