Green Synthesis of Silver Nanoparticles using Cinnamomum Zylinicum and their Synergistic Effect against Multi-Drug Resistance Bacteria
Article Information
Hamsa I Almalah, Hind A Alzahrani, Hayam S Abdelkader*
University of Jeddah, Microbiology Department, College of Sciences, Saudi Arabia
*Corresponding Authors: Hayam S Abdelkader, University of Jeddah, Microbiology Department, College of Sciences, Saudi Arabia
Received: 05 August 2019; Accepted: 16 August 2019; Published: 20 August 2019
Citation:
Hamsa I Almalah, Hind A Alzahrani, Hayam S Abdelkader. Green Synthesis of Silver Nanoparticles using Cinnamomum Zylinicum and their Synergistic Effect against Multi-Drug Resistance Bacteria. Journal of Nanotechnology Research 1 (2019): 095-107.
Share at FacebookAbstract
Multidrug - resistant (MDR) bacteria are considered life-threatening and need fast identification and antibiotic sensitivity testing to overcome this problem. In the current article, we highlighting the potential of developing silver Nanoparticles (AgNPs) using Cinnamomum zylinicum bark extracts as a promising approach. UV, SEM, TEM and FT-IR analysis were carried out to characterize the biosynthesized AgNPs. UV-visible spectroscopy showed the presence of characterized peek at (420 nm), TEM showed spherical shaped and monodispersed nanoparticles of size range 10 to 78.9 nm and FT-IR spectrum confirmed the presence of various functional groups in the biomolecules which serve as a capping agent for the nanoparticles. Biosynthesized (AgNPs) have been evaluated as an antibacterial against MDR gram-negative bacteria Acinetobacter baumanni, Klebsiella pneumoniae, and Pseudomonas aeruginosa strains and gram-positive bacteria Staphylococcus aureus. The results showed that obtained silver Nanoparticles are efficient in inhibiting both gram-positive and gram-negative bacteria when compared with antibiotic giving a zone of inhibition of 25 mm against S. aureus, 24 mm against K. pneumonia, and P. aeruginosa and 22 mm against A. baumanni respectively. Furthermore, the effectiveness of AgNPs against these test strains was assessed with multiple broad spectrum antibiotics. The results demonstrated that the incorporation of antibiotics with AgNPs has amazing antibacterial effects. The highest extent was observed with gentamycin against S. aureas, K. pneumonia, P. aeruginosa and A. baumanni, respectively. The minimum inhibitory concentrations (MICs) of AgNPs were also determined using microdilution assay. This study gives encouragement that
Keywords
AgNPs, Cinnamomum zylinicum, UV-Visible spectroscopy, TEM, FT-IR, MDR bacteria, MICs
AgNPs articles AgNPs Research articles AgNPs review articles AgNPs PubMed articles AgNPs PubMed Central articles AgNPs 2023 articles AgNPs 2024 articles AgNPs Scopus articles AgNPs impact factor journals AgNPs Scopus journals AgNPs PubMed journals AgNPs medical journals AgNPs free journals AgNPs best journals AgNPs top journals AgNPs free medical journals AgNPs famous journals AgNPs Google Scholar indexed journals Cinnamomum zylinicum articles Cinnamomum zylinicum Research articles Cinnamomum zylinicum review articles Cinnamomum zylinicum PubMed articles Cinnamomum zylinicum PubMed Central articles Cinnamomum zylinicum 2023 articles Cinnamomum zylinicum 2024 articles Cinnamomum zylinicum Scopus articles Cinnamomum zylinicum impact factor journals Cinnamomum zylinicum Scopus journals Cinnamomum zylinicum PubMed journals Cinnamomum zylinicum medical journals Cinnamomum zylinicum free journals Cinnamomum zylinicum best journals Cinnamomum zylinicum top journals Cinnamomum zylinicum free medical journals Cinnamomum zylinicum famous journals Cinnamomum zylinicum Google Scholar indexed journals UV-Visible spectroscopy articles UV-Visible spectroscopy Research articles UV-Visible spectroscopy review articles UV-Visible spectroscopy PubMed articles UV-Visible spectroscopy PubMed Central articles UV-Visible spectroscopy 2023 articles UV-Visible spectroscopy 2024 articles UV-Visible spectroscopy Scopus articles UV-Visible spectroscopy impact factor journals UV-Visible spectroscopy Scopus journals UV-Visible spectroscopy PubMed journals UV-Visible spectroscopy medical journals UV-Visible spectroscopy free journals UV-Visible spectroscopy best journals UV-Visible spectroscopy top journals UV-Visible spectroscopy free medical journals UV-Visible spectroscopy famous journals UV-Visible spectroscopy Google Scholar indexed journals TEM articles TEM Research articles TEM review articles TEM PubMed articles TEM PubMed Central articles TEM 2023 articles TEM 2024 articles TEM Scopus articles TEM impact factor journals TEM Scopus journals TEM PubMed journals TEM medical journals TEM free journals TEM best journals TEM top journals TEM free medical journals TEM famous journals TEM Google Scholar indexed journals FT-IR articles FT-IR Research articles FT-IR review articles FT-IR PubMed articles FT-IR PubMed Central articles FT-IR 2023 articles FT-IR 2024 articles FT-IR Scopus articles FT-IR impact factor journals FT-IR Scopus journals FT-IR PubMed journals FT-IR medical journals FT-IR free journals FT-IR best journals FT-IR top journals FT-IR free medical journals FT-IR famous journals FT-IR Google Scholar indexed journals MDR bacteria articles MDR bacteria Research articles MDR bacteria review articles MDR bacteria PubMed articles MDR bacteria PubMed Central articles MDR bacteria 2023 articles MDR bacteria 2024 articles MDR bacteria Scopus articles MDR bacteria impact factor journals MDR bacteria Scopus journals MDR bacteria PubMed journals MDR bacteria medical journals MDR bacteria free journals MDR bacteria best journals MDR bacteria top journals MDR bacteria free medical journals MDR bacteria famous journals MDR bacteria Google Scholar indexed journals MICs articles MICs Research articles MICs review articles MICs PubMed articles MICs PubMed Central articles MICs 2023 articles MICs 2024 articles MICs Scopus articles MICs impact factor journals MICs Scopus journals MICs PubMed journals MICs medical journals MICs free journals MICs best journals MICs top journals MICs free medical journals MICs famous journals MICs Google Scholar indexed journals K. pneumonia articles K. pneumonia Research articles K. pneumonia review articles K. pneumonia PubMed articles K. pneumonia PubMed Central articles K. pneumonia 2023 articles K. pneumonia 2024 articles K. pneumonia Scopus articles K. pneumonia impact factor journals K. pneumonia Scopus journals K. pneumonia PubMed journals K. pneumonia medical journals K. pneumonia free journals K. pneumonia best journals K. pneumonia top journals K. pneumonia free medical journals K. pneumonia famous journals K. pneumonia Google Scholar indexed journals antibiotic-resistant articles antibiotic-resistant Research articles antibiotic-resistant review articles antibiotic-resistant PubMed articles antibiotic-resistant PubMed Central articles antibiotic-resistant 2023 articles antibiotic-resistant 2024 articles antibiotic-resistant Scopus articles antibiotic-resistant impact factor journals antibiotic-resistant Scopus journals antibiotic-resistant PubMed journals antibiotic-resistant medical journals antibiotic-resistant free journals antibiotic-resistant best journals antibiotic-resistant top journals antibiotic-resistant free medical journals antibiotic-resistant famous journals antibiotic-resistant Google Scholar indexed journals microbes articles microbes Research articles microbes review articles microbes PubMed articles microbes PubMed Central articles microbes 2023 articles microbes 2024 articles microbes Scopus articles microbes impact factor journals microbes Scopus journals microbes PubMed journals microbes medical journals microbes free journals microbes best journals microbes top journals microbes free medical journals microbes famous journals microbes Google Scholar indexed journals
Article Details
1. Introduction
Nanotechnology is the potential future solution for the elimination of antibiotic-resistant microbes, which can stimulate the creation and synthesis of a new generation of antibiotics [1]. Nanotechnology has an important role in promoting the treatment of highly virulent bacterial diseases, especially those that resistant to antibiotics and are difficult to control with traditional treatments [2-6]. The antibiotic resistant bacteria has increased rapidly in the last few years, and become a growing problem that has impacted the world and brought about the beginning of the end for the old generation of antibiotics [7]. The miss-use of traditional antibiotics to combat bacterial infection has led to the current and major challenge of resistant bacteria, especially the multi-resistant bacteria (MDR) such as Methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumonia, and Neisseria meningitides [8]. It is known that silver is capable of killing 650 microbial germs that harm the human body [9]. Extensive studies have been carried out in evaluating antibacterial potentials of Nano particles. The antibacterial effects exhibited by NPs accentuate the potential of their advancement into future generation of antimicrobial agents [10]. NPs derived from plants that manifest promising antibacterial activities have high potential to be developed into future antibacterials mainly due to their low toxic effects [11].
NPs have been previously reported to inhibit gram-positive bacteria such as Staphylococcus spp [11, 12] Streptococcus spp. [13], and Bacillus spp [14, 15], and gram-negative bacteria such as Escherichia spp [16], Pseudomonas spp [17], Salmonella spp [18], Shigella spp [17, 21], Proteus spp. [21, 22], and Vibrio spp. [21, 23]. More promising, NPs have also shown potential to inhibit antibiotic-resistant bacteria such as Methicillin-resistant S. aureus (MRSA) [24, 25] and drug-resistant E. coli [26]. Previous studies have also manifested the diverse mechanisms for the silver Nanoparticles’ bactericidal effect. Besides that AgNPs interact with the surface of the membrane, they can also penetrate into the bacterial cell membrane [27]. Moreover, silver Nanoparticles can attach to the DNA inside the bacterial cells, suppressing its replication or interaction with the bacterial ribosome [28]. It has been observed that silver Nanoparticles have the ability to damage the structure of the bacterial cell membranous enzymes, which cause bacterial death eventually [29]. The goal of this study is to develop new therapeutic antibacterial drug using silver Nanoparticles to combat the growing threat of antibiotic-resistant bacteria.
2. Materials and Methods
2.1 Bacterial identification
Nutrient broth and Nutrient agar, Mueller Hinton Broth (MHB), Mueller Hinton Agar (MHA), silver nitrate, were purchased from Sigma-Aldrich (St. Louis, MO, USA). The Staphylococcus aureus, Acintetobacter baumanni and Klebsiella pneumoniae, and Pseudomonas aeruginosa mutans strains used in the present study were collected from King AbdulAziz hospital, Mekkah, KSA. All four strains were routinely grown in nutrient agar at 37°C. The characterization of bacteria was performed as outlined by Kalimuthu et al. [30]. The morphological characterization was carried out according to the methods indicated in Bergey’s Manual. Molecular characterization was conducted using the 16s rRNA technique. The nucleotide sequences of resistant strains of P. aeruginosa and K. pneumoni have been submitted to GenBank under accession no. MN121254, MN121255, MN121256, and MN121250 respectively.
2.2 Preparation of aqueous extract of C. zylinicum
10 gm of Barks of C. zylinicum were used for biosynthesis of the AgNPs due to its cost effectiveness, ease of availability and medicinal properties. C. zylinicum was homogenized into fine powder and boiled with 100 mL distilled water for 30 min at 60°C and the extract was obtained after filtration through Whatman No. 1 filter paper and was used for biosynthesis experiments [31].
2.3 Biosynthesis of AgNPs using C. zylinicum aqueous extracts
10 mL of aqueous extract of C. zylinicum was mixed with 1 mM of silver nitrate solution (100 mL) in the dark condition for 24 h. The synthesis of silver nanoparticles was done at 60°C by adding the aqueous extract of C. zylinicum drop by drop on magnetic stirrer. A change in the color of the solution from light yellow to dark brown indicated the synthesis of AgNPs. The obtained yield of AgNPs was calculated according to Kalishwaralal et al. [32].
2.4 Characterization of synthesized silver nanoparticles
The biosynthesized AgNPs were characterized according to the method described by Gurunathan et al. [33]. UV-vis spectra were measured using a (UV-2450, Shimadzu, Tokyo, Japan). The shape and size of silver nanoparticles were determined by TEM. For TEM, a drop of aqueous silver nanoparticles sample was fixed on a carbon coated grid, and let dry in room temperature; the micrographs were acquired using TEM. The particle sizes were measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS90 (Malvern Instruments, Malvern, UK). Fourier transform infrared spectroscopy (FT-IR) (Thermo Scientific Smart iTR™ ) was used to characterize the changes and the composition on the surface of the synthesized nanoparticles.
2.5 Determination of the minimum inhibitory concentrations (MICs) of AgNPs
Two-fold broth microdilution of AgNPs and different antibiotics were carried out in 96-well microtiter plates using MHB according to the Clinical and Laboratory Standards Institute (CLSI 2005). To examine AgNP MICs, S. aureus, A. baumanni and K. neumoniae, and P. aeruginosa mutans strains were exposed to 0-10 µg/mL AgNPs prepared in phosphate-buffered saline (PBS). Each concentration of AgNP solution was mixed with 1 mL of the bacterial suspension in MH media until the final concentration of bacteria is ~105-106 (CFUs)/mL O/N. Then, 10-fold dilution has been done and 100 µL of which was cultured on MH agar media. The viability loss was estimated by colony counting method and compared with those on control plates (AgNP free MH media). All experiments were performed independently in triplicate. The MIC was defined as the minimum concentration of AgNPs that inhibit the growth of bacteria. Control tests were carried out with solutions containing all the reaction components except for AgNPs. In addition, different concentrations of antibiotics alone, AgNPs alone, and/or a combination of both were carried out in different microtitre plates under the same conditions. The optical density of each well was measured at 600 nm by using ELISA reader. All samples were repeated trice and the average values were calculated independently.
2.6 Agar diffusion assay
The agar diffusion assay was accomplished as illustrated by Shahverdi et al. The bactericidal activity of antibiotics in combination with or without AgNPs was tested against bacterial strains on Mueller Hinton Agar plates. Broad-spectrum antibiotics were selected to analyze the combined effect of antibiotics and AgNPs. According to CLSI standard, antibiotics were used at appropriate concentrations as follows: amoxycillin (10 µg/mL), cefatoxime (25 µg/mL), sulbactam (15 µg/mL), gentamicin (20 µg/mL), and flucoxacilin (30 µg/mL). Each filter disk was soaked with the minimum inhibitory concentration of AgNPs for each bacterial strain. A single colony of each test strain was grown overnight in MHB on shaker with (200 rpm) at 37°C. 0.5 McFarland standard was composed by diluting the overnight bacterial cultures with 0.9% NaCl and then plated on MH agar plates together with the prepared disks containing several antibiotics. Similar experiments were executed with AgNPs alone. After incubation at 37°C for 24 h, a zone of inhibition (ZOI) was measured. The assays were completed in triplicate. The increment in antibacterial activity of different antibiotics has been calculated by the formula (B - A)/A x 100, where A and B are the ZOI for antibiotic and antibiotic+AgNPs, respectively [34].
2.7 Disk diffusion assay to evaluate synergistic effect
To evaluate the synergistic effects of green synthesized silver nanoparticles with various antibiotics were studied according to [35] against MDR strain on MH agar plates using disk diffusion method. The antibiotic discs used were amoxycillin (10 µg/mL), cefatoxime (25 µg/mL), sulbactam (15 µg/mL), gentamicin (20 µg/mL), and flucoxacilin (30 µg/mL). The bacterial inoculum was diluted in 5 ml of NaCl to obtain 0.5McFarland and cultured on the plate. Each disc was soaked in 5µL of different concentration (10, 20, 40, 60, 80, and 100) µg/ml of AgNPs, and then the plates were incubated at 37°C overnight. The inhibition zones were measured as described by Fayaz et al. [35] and compared with those obtained from antibiotics which tested in Antibiotic susceptibility. Statistical analysis was performed using all the assays were performed in triplicate and the results are presented as means ± SD.
3. Results and Discussion
3.1 UV-Vis spectroscopy
Addition of bark extract of C. zylinicum into the aqueous solution of silver nitrate produced a color change in the mixture to brown (Figure 3) duration the reaction period due to silver nanoparticles’ Surface plasmon resonance (SPR) excitation [36]. When 5 ml of bark extract of C. zylinicum was added to1 mM of silver nitrate solution (10 ml), the color of the solution changed from faint light to colloidal brown suggesting the formation of silver nanoparticles. UV spectra of Plasmon resonance band was observed at 420 (Figure 4) according to those reported by [37]. The UV-Visible spectra recorded after 1 h from the initiation of reaction. The results of the UV-vis spectra recorded, suggested that the bioreduction was achieved using C. zylinicum bark extract as reducing agent and that formation of silver nanoparticles occurred rapidly within 1h. These results were in agreement with that reported by Ahmed et al. who found that A. indica leaf extract was the most rapid reducing agent for silver nitrate solution and the UV-vis spectra was detected rapidly within 15 min indicating the formation of silver nanoparticles.
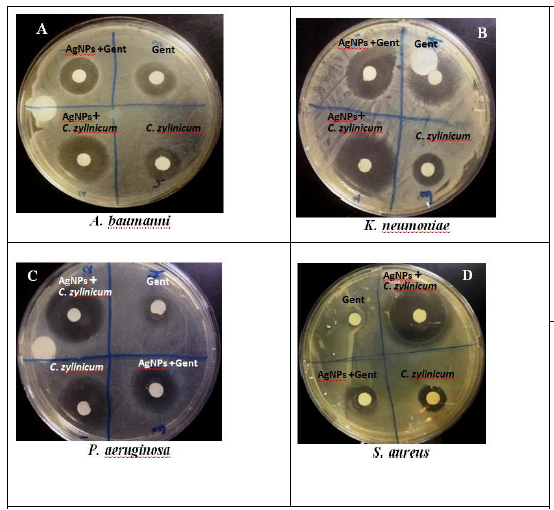
Figure 1: a Zone of inhibition (ZOI) graph of AgNPs/C. zylinicum, AgNPs/Gent, C. zylinicum extract, and Gentamycin against (A): A. baumanni, (B): K. neumoniae, (C): P. aeruginosa, and (D): S. aureus.
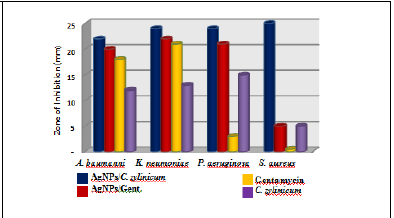
Figure 2: Photograph of zone of inhibition of AgNPs/C.zylinicum, and AgNPs/Gentamycin, Gentamycin, and C. zylinicum barkextract against (A) A. baumanni , (B) K. pneumoniae, (C) P. aeruginosa, (D) S. aureus.
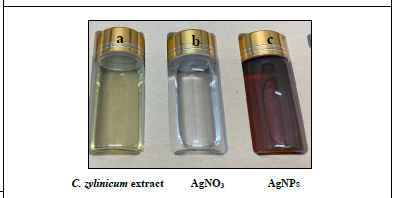
Figure 3: (a) Light yellow color of the C.zylinicum bark extract, (b) 1 mM of AgNO3 solution, (c) dark brown color of the reaction mixture indicating the formation of AgNPs.
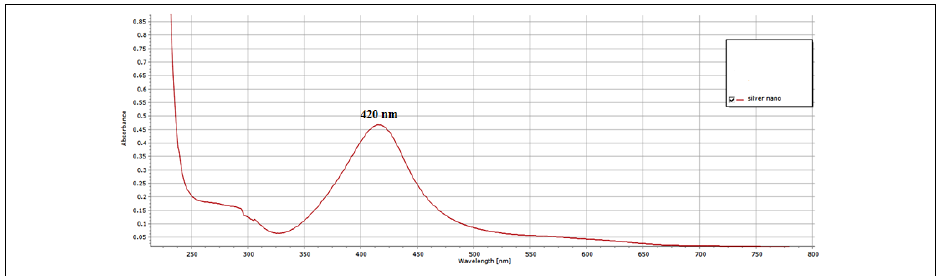
Figure 4: UV-visible absorption spectrum of biosynthesized AgNPs by the reduction of AgNO3 solution with the C. zyilinicum after 1h.
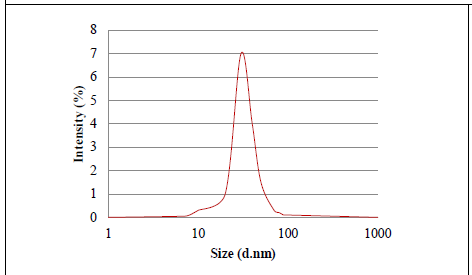
Figure 5: Graph representing the distribution particle size of biosynthesized AgNPs.
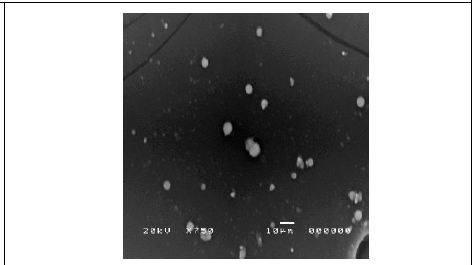
Figure 6: SEM micrograph of synthesized silver nanoparticles using C. zylinicum bark extract magnified 750x.
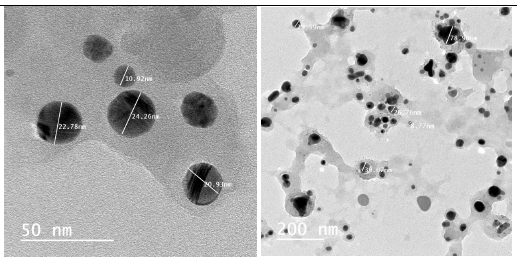
Figure 7:Transmission electron microscopic (TEM) images of the biosynthesized AgNPs.
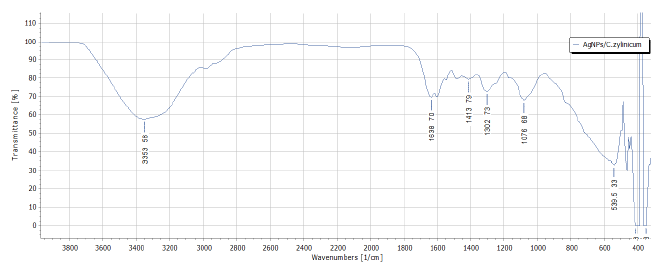
Figure 8:FTIR analysis of biosynthesized AgNPs/C.zylinicum.
|
Tested strains |
Gentamycine (Standard antibacterial agent) |
*Diameter of inhibition zone (mm) produced by AgNPs (Mean ± SD) |
*Diameter of inhibition zone (mm) produced by AgNPs + antibiotic (Mean ± SD) |
MICs of AgNPs (µg/ml) |
p-value |
|
Staphylococcus aureus |
0.46 ± 0.06 |
25 ± 0.20 |
4.8 ± 0.2 |
4.5 ± 0.04 |
0 .096 |
|
Acinetobacter baumannii |
17.3 ± 0.48 |
22 ± 0.21 |
19.9 ± 0.4 |
5.7 ± 0.02 |
0.057 |
|
Pseudomonas aeruginosa |
2.8 ± 0.20 |
24 ± 0.14 |
20.8 ± 0.3 |
3.1 ± 0.01 |
0.072 |
|
Klebsiella pneumonia |
20.7 ± 0.32 |
24 ± 0.19 |
21.5 ± 0.6 |
2.8 ± 0.03 |
0.01 |
|
*Three repeats were performed for each tested strain p-value significant <0.05 p-value non-significant >0.05 AgNPs (20 µg/ml ), concentration of antibiotics (20 μg/ ml) |
|||||
Table 1: The antimicrobial activity (inhibition zone in mm) and the minimum inhibitory concentration values of the biosynthesized AgNPs against different bacterial strains.
3.2 SEM, TEM and particle size
The dynamic light scattering (DLS) profile of the synthesized AgNPs using C. zylinicum aqueous bark extract is shown in (Figure 5). The shape of the synthesized silver nanoparticles was analyzed by SEM magnified at 750x is shown in (Figure 6). The image obtained by the SEM showed spherical nanoparticles and some particles of irregular shape. As well as, the biosynthesized silver nanoparticles were studied by TEM. The image obviously showed that particles size were in the range from 10 to 78.9 nm (Figure 7). The nanoparticles are spherical shaped and homogeneous which comply with the shape of SPR band in the UV-visible spectrum.
3.3 FTIR analysis of AgNPs
FTIR analysis of silver nanoparticles was conducted to confirm the role of the plant extract as a capping and reducing agent and existence of specific chemical groups responsible for the characteristic chemical reactions of biosynthesized AgNPs (Figure 8). The broad band at 3353 cm-1 is due to stretching vibration of bonded and non-bonded –O–H groups. The band at 1638 cm-1 are attributable to stretching vibration of carbonyl group which is characteristic for the secondary amides and other compounds containing C=O group [38]. The strong bands at 1413 cm–1 correspond to the bending vibrations of CH which is indicative for the lignins presence. The observed peak at 1302 denote C(O)–O stretching vibrations and –OH in plane vibrations/amide III (e.g. in aromatic ethers). The absorption bands at 1100-1000 cm–1 in the fingerprint region suggest several modes such as C-H deformation or C-O or C-C stretching, pertaining to carbohydrates. The C-O-C groups exhibit strong bands at 1076 cm–1 [39]. The absorbance bands at 539 cm–1 represent C-O-O and P-O-C bending of aromatic compounds (phosphates). The phytochemical constituents of C. zylinicum bark extract are expected to interact with metal salts through these functional groups and facilitate their reduction to nanoparticles [40].
3.4 Antimicrobial activity of silver nanoparticles
Silver nanoparticles exhibiting antimicrobial activity against multidrug resistant bacteria, with different extents, as showed by the diameter of inhibition zone (Figure 1(A)-(B)-(C). The Gram negative bacteria (A. baumanni, K. pneumoniae, and P. aeruginosa) strains exhibited larger inhibition zones, compared with the Gram positive bacteria (S. aureus) (Figure 1D), that may vary because of the structure of the cell wall. The composition of Gram- positive cell wall contained a dense peptidoglycan layer composed of linear polyaccharide chains crossed through brief peptides forming a stiffer framework that makes silver nanoparticles hard to penetrate while there is a thin layer of peptidoglycan in Gram negative bacteria the cell wall [41]. The results of antibacterial activities of biosynthesized silver nanoparticles evaluated from the agar well diffusion method are given in (Figure 1). C. zylinica extract mediated silver nanoparticles revealed that the AgNPs showed great antibacterial activity against both gram negative and gram positive bacterial strain where the inhibition zone diameter were (25, 22, 24, and 24 mm) for S. aureus, A. baumannii, P. aeruginosa, and K. pneumonia, respectively (Table 1). However, gentamycine and C. zylinicum bark extract alone showed very low antibacterial activity because of the water medium of extraction as well as lower concentration used in the experiment. Although, it is to be presumed that, the tested bacterial strains were supposed to be resistant for gentamycine, it shows very low activity against A. baumanni, K. neumoniae, and P. aeruginosa, while it has no activity against S. aureus. Based on inhibition zone assay, biosynthesized silver nanoparticles combined with gentamycine antibiotic showed a great synergistic antibacterial activity against all tested bacterial strains (Figure 1).
MIC was used to analyze the antimicrobial activity of silver nanoparticles. The technique which used to detect the MIC of silver nanoparticles is broth microdilution methods. The MIC is defined as the lowest concentration of the antimicrobial compound that inhibits the growth of a microorganism [42]. After 24 h of incubation at 37°C, no growth of A. baumanni, K. pneumoniae, and P. aeruginosa, and S. aureus in the microtitre plate supplemented with 5.7, 2.8, 3.1 and 4.5 µg/ml of silver nanoparticles was observed, and the optical density was 0.041, 0.017, 0.038 and 0.021, respectively. Therefore, the MICs were 5.7, 2.8, 3.1 and 4.5 µg/ml, respectively (Table 1). The biological characteristics of silver nanoparticles against microorganisms are underpinned by several mechanisms. Firstly, silver nanoparticles bind to negatively charged cell surfaces, alter cell membrane and cell wall physical and chemical characteristics and interfere with key functions such as permeability, osmoregulation, electron transport and respiration [27, 43, 44, 45]. Second, silver nanoparticles can interact with DNA, thiol group of L-cysteine protein and other phosphorus- and sulfur-containing cell constituents causing enzymatic dysfunction [43, 45, 46, 47]. Finally, the silver nanoparticles cause damage on proteins and DNA via release of reactive oxygen species (ROS) [48]. Third, Silver nanoparticles release silver ions, which generate a size and dose-dependent enhanced biocidal impact [45, 49].
4. Conclusions
Green synthesis of silver nanoparticles has a promising antibacterial action and has a great synergistic effect in enhancing the efficacy of antibiotics. There is now a huge effort in the production of extremely powerful and robust NPs for clinical use.
References
- Wang L, Hu C and Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine 12 (2017): 1227-1249.
- Aderibigbe BA. Metal-Based Nanoparticles for the Treatment of Infectious Diseases. Molecules 22 (2017).
- AlMatar M, Makky EA, Var I, et al. The role of nanoparticles in the inhibition of multidrug-resistant bacteria and biofilms. Curr Drug Deliv 15 (2017): 470-484.
- Hemeg HA. Nanomaterials for alternative antibacterial therapy. Int J Nanomed 12 (2017): 8211-8225.
- Bassegoda A, Ivanova K, Ramon E, et al. Strategies to prevent the occurrence of resistance against antibiotics by using advanced materials. Appl Microbiol Biotechnol 102 (2018): 2075-2089.
- Siddiqi KS, Husen A and Rao RAK. A review on biosynthesis of silver nanoparticles and their biocidal properties. J Nanobiotechnology 16 (2018): 14.
- Fair RJ and Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem 6 (2014): 25-64.
- Barker KF. Antibiotic resistance: a current perspective. Br J Clin Pharmacol 48 (1999): 109-124.
- Haytham MM Ibrahim. Green synthesis and characterization of silver nanoparticles using banana peel extract and their antimicrobial activity against representative microorganisms. J Radiation Res Appl Sci 8 (2015): 265.
- Teow SY, Wong M, Yap HY, et al. Bactericidal Properties of Plants-Derived Metal and Metal Oxide Nanoparticles (NPs). Molecules 23 (2018): 1366.
- Gupta A, Eral HB, Hatton TA, et al. Nanoemulsions: Formation, properties and applications. Soft Matter 12 (2016): 2826-2841.
- Jiménez Pérez ZE, Mathiyalagan R, Markus J, et al. Ginseng-berry-mediated gold and silver nanoparticle synthesis and evaluation of their in vitro antioxidant, antimicrobial, and cytotoxicity effects on human dermal ?broblast and murine melanoma skin cell lines. Int J Nanomed 12 (2017): 709-723.
- Kelkawi AHA, Abbasi Kajani A and Bordbar AK. Green synthesis of silver nanoparticles using Menthapulegium and investigation of their antibacterial, antifungal and anticancer activity. IET Nanobiotechnol 11 (2017): 370-376.
- Dehghanizade S, Arasteh J, Mirzaie A. Green synthesis of silver nanoparticles using Anthemis atropatana extract: Characterization and in vitro biological activities. Artif Cell Nanomed Biotechnol 46 (2018): 160-168.
- Manikandan V, Velmurugan P, Park JH, et al. Green synthesis of silver oxide nanoparticles and its antibacterial activity against dental pathogens. 3 Biotech 7 (2017): 72.
- Skandalis N, Dimopoulou A, Georgopoulou A, et al. The effect of silver nanoparticles size, produced using plant extract from Arbutus unedo on their antibacterial efficacy. Nanomaterials 7 (2017): E178.
- Patra JK and Baek KH. Antibacterial activity and synergistic antibacterial potential of biosynthesized silver nanoparticles against foodborne pathogenic bacteria along with its anticandidal and antioxidant effects. Front. Microbiol 8 (2017): 167.
- Naraginti S and Li Y. Preliminary investigation of catalytic, antioxidant, anticancer and bactericidal activity of green synthesized silver and gold nanoparticles using Actinidia deliciosa. J Photochem Photobiol B Biol 170 (2017): 225-234.
- Kanjikar AP, Hugar AL and Londonkar RL. Characterization of phyto-nanoparticles from Ficus krishnae for their antibacterial and anticancer activities. Drug Dev Ind Pharm 44 (2018): 377-384.
- Zia M, Gul S, Akhtar J, et al. Green synthesis of silver nanoparticles from grape and tomato juices and evaluation of biological activities. IET Nanobiotechnol 11 (2017): 193-199.
- Banumathi B, Vaseeharan B, Ishwarya R, et al. Toxicity of herbal extracts used in ethno-veterinary medicine and green-encapsulated ZnO nanoparticles against Aedes aegypti and microbial pathogens. Parasitol Res 116 (2017): 1637-1651.
- Rajakumar G, Gomathi T, Thiruvengadam M, et al. Evaluation of anti-cholinesterase, antibacterial and cytotoxic activities of green synthesized silver nanoparticles using from Millettia pinnata flower extract. Microb Pathog 103 (2017): 123-128.
- Kumar M, Bala R, Gondil VS, et al. Efficient, Green and One Pot Synthesis of Sodium Acetate Functionalized Silver Nanoparticles and Their Potential Application as Food Preservative. BioNanoSci 7 (2017): 521-529.
- Jadhav K, Dhamecha D, Bhattacharya D, et al. Green and ecofriendly synthesis of silver nanoparticles: Characterization, biocompatibility studies and gel formulation for treatment of infections in burns. J Photochem Photobiol B Biol 155 (2016): 109-115.
- Azizi S, Mohamad R, Mahdavi Shahri M. Green microwave-assisted combustion synthesis of zinc oxide nanoparticles with Citrullus colocynthis L. schrad: Characterization and biomedical applications. Molecules 22 (2017): 301.
- Khan FU, Chen Y, Khan NU, et al. Visible light inactivation of E. coli, cytotoxicity and ROS determination of biochemically capped gold nanoparticles. Microb Pathog 107 (2017): 419-424.
- Sondi I and Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. Journal of Colloid and Interface Science 275 (2004): 177-182.
- Lei Z, Mingyu S, Xiao W, et al. Antioxidant stress is promoted by nano-anatase in spinach chloroplasts under UV-B radiation. Biol Trace Elem Res 121 (2008): 69-79.
- Wen-Ru L, Xiao-Bao X, Qing-Shan S, et al. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biotechnol 85 (2010): 1115-1122.
- Kalimuthu K, Babu RS, Venkataraman D, et al. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf B 65 (2008): 150-153.
- Lekshmi JP, Sumi SB, Viveka S, et al. Antibacterial activity of nanoparticles from Allium sp. J Microbiol Biotechnol Res 2 (2012): 115-119.
- Kalishwaralal K, BarathManiKanth S, Pandian SR, et al. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf B Biointerfaces 79 (2010): 340-344.
- Gurunathan S, Han JW, Kwon DN, et al. Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale Res Lett 9 (2014): 373.
- Fayaz AM, Balaji K, Girilal M, et al. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram negative bacteria. Nanomedicine: Nanotechnology, Biology, and Medicine 61 (2010): 103-109.
- Mahmood MA. The antibacterial effect of silver nanoparticles on some bacterial pathogens. Iraqi Journal of Physics 3 (2012): 697-708.
- Veerasamy R, Xin TZ, Gunasagaran S, et al. Biosynthesis of silver nanoparticles using mangosteen leaf extract and evaluation of their antimicrobial activities. Journal of Saudi Chemical Society 15 (2011): 113-120.
- Obaid AY, Al-Thabaiti SA, Al-Harbi LM, et al. Extracellular bio-synthesis of silver nanoparticles. Global Advanced Research Journal of Microbiology 3 (2015): 119-126.
- Mueen Ahmed KK, Rana AC and Dixit VK. Calotropis species (Asclepediaceae). A comprehensive review. Pharmacognosy Magazine 1 (2005): 48-52.
- Li YM, Sun SQ, Zhou Q, et al. Identification of American ginseng from different regions using FT-IR and two-dimensional correlation IR spectroscopy. Vibrational spectroscopy 36 (2004): 227-232.
- Bar H, Bhui DK, Sahoo GP, et al. Green synthesis of silver nanoparticles using seed extract of Jatrophacurcas. Colloids and Surfaces A-Physicochemical and Engineering Aspects 348 (2009): 212-216.
- Shrivastava S, Bera T, Roy A, et al. Characterization of enhanced antibacterial effects of novel silver nanoparticles. Nanotechnology 18 (2007): 103-112.
- Wiegand I, Hilpert K and Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols 3 (2008): 163-175.
- Nel AE, Madler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nature Materials 8 (2009): 543-557.
- Su HL, Chou CC, Hung DJ, et al. The disruption of bacterial membrane integrity through ROS generation induced by nanohybrids of silver and clay. Biomaterials 30 (2009): 5979-5987.
- Marambio-Jones C and Hoek EMV. A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. Journal of Nanoparticle Research 12 (2010): 1531-1551.
- AshaRani PV, Mun GLK, Hande MP, et al. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 3 (2009): 279-290.
- Gordon O, Vig Slenters T, Brunetto PS, et al. Silver coordination polymers for prevention of implant infection: thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction. Antimicrob Agents Chemother 54 (2010): 4208-4218.
- Hossain Z and Huq F. Studies on the interaction between Ag(+) and DNA. J Inorg Biochem 91 (2002): 398-404.
- Liu JY, Sonshine DA, Shervani S, et al. Controlled release of biologically active silver from nanosilver surfaces. ACS Nano 4 (2010): 6903-6913.
