Genome-Wide Cnv Study and Functional Evaluation Identified Ctdspl as Tumour Suppressor Gene for Cervical Cancer
Article Information
Dandan Zhang1,2,?,Shuai Wang3,?, Zhenli Li4,?, Tao Cui5,?, Xiunian Chen1,2, Jingwen Gong1,2, Yatao Du6, Yuexin Gan6, Xiaoguang Ren7, Jinyan Huang8, Patrik KE Magnusson9, Ping Zhang10, Xingping Zhao11, Dabao Xu11, Wenqiang Yu7, Huibo Wang3,12*, Gyllensten Ulf5*, Dan Chen11,13*
1Department of Pathology, and Department of Medical Oncology of the Second Affiliated Hospital,Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310058, China
2Department of Pathology, Key Laboratory of Disease Proteomics of Zhejiang Province, School of Medicine, Zhejiang University, Hangzhou, 310058, China
3Department of Hematology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China
4The United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou 350025, P. R. China
5Department of Immunology, Genetics and Pathology, Science for Life Laboratory Uppsala, Uppsala University, Uppsala, Sweden
6Ministry of Education and Shanghai Key Laboratory of Children’s Environmental Health, Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
7Laboratory of RNA Epigenetics, Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai,R. China.
8Biomedical big data center, the First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
9Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm 17177, Sweden
10Department of Gynecology, Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
11Department of Gynecology, Third Xiangya Hospital of Central South University, Changsha, China. 12Department of Neurosurgery, First Affiliated Hospital of Nanjing Medical University, Nanjing, China 13Shanghai Fosun Pharmaceutical Industrial Development, Co., Ltd. Shanghai, China
*Corresponding Authors: Dan Chen, Department of Gynecology, Third Xiangya Hospital of Central South University, Changsha, China and Shanghai Fosun Pharmaceutical Industrial Development, Co., Ltd. Shanghai, China. Ulf Gyllensten, Department of Immunology, Genetics and Pathology, Science for Life Laboratory Uppsala, Uppsala University, Uppsala, Sweden. Huibo Wang, Department of Hematology, First Affiliated Hospital of Nanjing Medical University, Nanjing, China and Department of Neurosurgery, First Affiliated Hospital of Nanjing Medical University, Nanjing, China.
?These authors contributed equally
Received: 06 September 2022; Accepted: 12 September 2022; Published: 27 October 2022
Citation:
Zhang et al. Genome-wide CNV study and functional evaluation identified CTDSPL as tumour suppressor gene for cervical cancer. Obstetrics and Gynecology Research 5 (2022): 261-273
Share at FacebookAbstract
Background: Copy number variations (CNVs) may explain some of the missing heritability not identified in genome-wide association studies (GWASs).
Method: We performed the first genome-wide study of both common and rare germline CNVs in relation to cervical cancer by analyzing 731,422 single-nucleotide polymorphisms (SNPs) in 1,034 cervical cancer cases and 3,948 controls, followed by replication in 1,396 cases and 1,057 controls.
Results: We found that a 6367bp deletion in intron 1 of the CTD small phosphatase like gene (CTDSPL) was associated with 2.54- fold increased risk of cervical cancer (odds ratio = 2.54, 95% confidence interval =2.08-3.12, P = 2.0×10-19). This CNV is one of the strongest common genetic risk variants identified so far for cervical cancer. The deletion removes the binding sites of zinc finger protein 263 (ZNF263), binding protein 2 (GATA2) and interferon regulatory factor 1(IRF1), and hence downregulates the transcription of CTDSPL. HeLa cells expressing CTDSPL showed a significant decrease in colony- forming ability. Compared with control groups, mice injected with HeLa cells expressing CTDSPL exhibited a significant reduction in tumour volume. Furthermore, CTDSPL-depleted immortalized End1/E6E7 could form tumours in NOD- SCID mice.
Conclusion: These findings indicate that CTDSPL is a tumour suppressor gene for cervical cancer and the 6367bp deletion downregulates CTDSPL transcription by removing binding sites of ZNF263, GATA2 and IRF1.
Keywords
Cervical cancer; Genome-wide association study; Copy number variants; CTD small phosphatase like gene; Tumour suppressor; Deletion
Cervical cancer articles Cervical cancer Research articles Cervical cancer review articles Cervical cancer PubMed articles Cervical cancer PubMed Central articles Cervical cancer 2023 articles Cervical cancer 2024 articles Cervical cancer Scopus articles Cervical cancer impact factor journals Cervical cancer Scopus journals Cervical cancer PubMed journals Cervical cancer medical journals Cervical cancer free journals Cervical cancer best journals Cervical cancer top journals Cervical cancer free medical journals Cervical cancer famous journals Cervical cancer Google Scholar indexed journals Genome-wide association study articles Genome-wide association study Research articles Genome-wide association study review articles Genome-wide association study PubMed articles Genome-wide association study PubMed Central articles Genome-wide association study 2023 articles Genome-wide association study 2024 articles Genome-wide association study Scopus articles Genome-wide association study impact factor journals Genome-wide association study Scopus journals Genome-wide association study PubMed journals Genome-wide association study medical journals Genome-wide association study free journals Genome-wide association study best journals Genome-wide association study top journals Genome-wide association study free medical journals Genome-wide association study famous journals Genome-wide association study Google Scholar indexed journals Copy number variants articles Copy number variants Research articles Copy number variants review articles Copy number variants PubMed articles Copy number variants PubMed Central articles Copy number variants 2023 articles Copy number variants 2024 articles Copy number variants Scopus articles Copy number variants impact factor journals Copy number variants Scopus journals Copy number variants PubMed journals Copy number variants medical journals Copy number variants free journals Copy number variants best journals Copy number variants top journals Copy number variants free medical journals Copy number variants famous journals Copy number variants Google Scholar indexed journals CTD small phosphatase like gene articles CTD small phosphatase like gene Research articles CTD small phosphatase like gene review articles CTD small phosphatase like gene PubMed articles CTD small phosphatase like gene PubMed Central articles CTD small phosphatase like gene 2023 articles CTD small phosphatase like gene 2024 articles CTD small phosphatase like gene Scopus articles CTD small phosphatase like gene impact factor journals CTD small phosphatase like gene Scopus journals CTD small phosphatase like gene PubMed journals CTD small phosphatase like gene medical journals CTD small phosphatase like gene free journals CTD small phosphatase like gene best journals CTD small phosphatase like gene top journals CTD small phosphatase like gene free medical journals CTD small phosphatase like gene famous journals CTD small phosphatase like gene Google Scholar indexed journals Tumour suppressor articles Tumour suppressor Research articles Tumour suppressor review articles Tumour suppressor PubMed articles Tumour suppressor PubMed Central articles Tumour suppressor 2023 articles Tumour suppressor 2024 articles Tumour suppressor Scopus articles Tumour suppressor impact factor journals Tumour suppressor Scopus journals Tumour suppressor PubMed journals Tumour suppressor medical journals Tumour suppressor free journals Tumour suppressor best journals Tumour suppressor top journals Tumour suppressor free medical journals Tumour suppressor famous journals Tumour suppressor Google Scholar indexed journals Deletion articles Deletion Research articles Deletion review articles Deletion PubMed articles Deletion PubMed Central articles Deletion 2023 articles Deletion 2024 articles Deletion Scopus articles Deletion impact factor journals Deletion Scopus journals Deletion PubMed journals Deletion medical journals Deletion free journals Deletion best journals Deletion top journals Deletion free medical journals Deletion famous journals Deletion Google Scholar indexed journals single-nucleotide polymorphisms articles single-nucleotide polymorphisms Research articles single-nucleotide polymorphisms review articles single-nucleotide polymorphisms PubMed articles single-nucleotide polymorphisms PubMed Central articles single-nucleotide polymorphisms 2023 articles single-nucleotide polymorphisms 2024 articles single-nucleotide polymorphisms Scopus articles single-nucleotide polymorphisms impact factor journals single-nucleotide polymorphisms Scopus journals single-nucleotide polymorphisms PubMed journals single-nucleotide polymorphisms medical journals single-nucleotide polymorphisms free journals single-nucleotide polymorphisms best journals single-nucleotide polymorphisms top journals single-nucleotide polymorphisms free medical journals single-nucleotide polymorphisms famous journals single-nucleotide polymorphisms Google Scholar indexed journals Cervix articles Cervix Research articles Cervix review articles Cervix PubMed articles Cervix PubMed Central articles Cervix 2023 articles Cervix 2024 articles Cervix Scopus articles Cervix impact factor journals Cervix Scopus journals Cervix PubMed journals Cervix medical journals Cervix free journals Cervix best journals Cervix top journals Cervix free medical journals Cervix famous journals Cervix Google Scholar indexed journals twins born articles twins born Research articles twins born review articles twins born PubMed articles twins born PubMed Central articles twins born 2023 articles twins born 2024 articles twins born Scopus articles twins born impact factor journals twins born Scopus journals twins born PubMed journals twins born medical journals twins born free journals twins born best journals twins born top journals twins born free medical journals twins born famous journals twins born Google Scholar indexed journals cervical squamous cell articles cervical squamous cell Research articles cervical squamous cell review articles cervical squamous cell PubMed articles cervical squamous cell PubMed Central articles cervical squamous cell 2023 articles cervical squamous cell 2024 articles cervical squamous cell Scopus articles cervical squamous cell impact factor journals cervical squamous cell Scopus journals cervical squamous cell PubMed journals cervical squamous cell medical journals cervical squamous cell free journals cervical squamous cell best journals cervical squamous cell top journals cervical squamous cell free medical journals cervical squamous cell famous journals cervical squamous cell Google Scholar indexed journals
Article Details
Background
Cervical cancer is the fourth most common cancer in women worldwide [1]. Genome-wide association studies (GWASs) focusing on single-nucleotide polymorphisms (SNPs) have identified multiple genetic susceptibility loci for cervical cancer [2, 3]. However, the risk variants identified to date have small effect sizes (per allele odds ratio [OR] < 1.50) and only explain a small fraction of the heritability. Although epistatic and gene-environment interactions may contribute to the unexplained heritability of cervical cancer, it seems likely that a significant fraction is due to loci that have not yet been identified. Recent studies indicate that copy number variations (CNVs) occur frequently in the genome and are an important source of human genetic variation [4, 5]. It has been proposed that CNVs may explain some of the missing heritability for complex diseases after the findings from GWASs [6]. The role of somatic CNVs in cervical cancer has been extensively studied [7-13]. However, very few studies have evaluated the association of germline CNVs with cervical cancer risk. Only one study demonstrated that a lower defensin beta 104A (DEFB4) copy number was associated with susceptibility to cervical cancer [14]. To assess the association of both common and rare germline CNVs with cervical cancer risk, we conducted a genome-wide CNV study of 1,034 cervical cases and 3,948 controls in a Swedish population using Illumina HumanOmniExpress BeadChip (Illumina, San Diego, CA) (731,422 SNPs) followed by a replication study of 1,396 cervical cancer subjects and 1,057 controls. We revealed that a 6367bp deletion in intron 1 of CTDSPL increased the risk of cervical cancer. We further explored the functions of CTDSPL in cervical cancers in vitro and in vivo.
Methods
Study population
Subjects included in the discovery phase were from a GWAS of cervical cancer in the Swedish population. The details of population and quality controls have been described elsewhere [2]. Briefly, subjects included in the discovery phase were from two studies, the CervixCan I study and the TwinGene study. CervixCan study included two parts, i.e. CervixCan Ι study that comprised cases who are the sole participants of their family and CervixCan II study that comprised individuals with more than one first-degree relative also participating. 766 sole participants (720 CIS and 46 invasive carcinoma) from the CervixCan Ι study were included in the discovery phase. The TwinGene study was a population-based Swedish study of twins born between 1911 and 1958. In total, 9,896 subjects were genotyped consecutively with those from the CervixCan Ι study using Illumina HumanOmniExpress BeadChip (731,422 SNPs) at the SNP&SEQ Technology Platform Uppsala, Sweden. Among these subjects, 309 unrelated cervical cancer cases (288 CIS and 21 invasive carcinoma) were further included in the discovery phase. One female singleton was then randomly selected from each twin pair without cervical cancer, resulting in 4,014 unrelated cervical cancer-free females who were included as controls. After stringent quality control [2], there were genotyping data in the discovery phase including 632,668 SNPs with an overall call rate of 99.92 % in 1,034 cervical cancer patients (971 with carcinoma in situ [CIS] and 63 with invasive carcinoma) and 3,948 control subjects. The replication series comprised 1,396 cervical cancer patients (1,265 CIS and 131 invasive carcinoma) from CervixCan II study and 1,057 controls. All the subjects were of Swedish descent. Informed consent was obtained from all subjects and each study was approved by the regional ethical review board in Uppsala, Sweden.
CNV detection
CNV coordinates were identified using both Penncnv and Quantisnp [15, 16]. Both of the two algorithms are based on a Hidden Markov Model (HMM), using intensity files generated by GenomeStudio software from Illumina. QuantiSNP2.0 is based on an objective Bayes HMM and takes into consideration log R Ratio (LRR) as well as B allele frequency (BAF) of each SNP. The PennCNV algorithm incorporates additional information including the population frequency of the B allele (PFB) and the distance between adjacent SNPs. To reduce false positive calls due to genomic waves, GC-content adjustment was performed to correct for the bias in both analyses [17]. The default setting was used for both algorithms.
Quality control
The initial sample quality control has been described elsewhere [2]. All the SNPs that pass quality control were included in CNV calling. The proportion of the array that was informative for the CNV calling was 632,668/731,422 = 86.5%. Samples were further filtered based on three additional criteria: individuals with more than 40 CNVs; an absolute value of GC wave factor (GCWF) larger than 0.02 or a standard deviation of LRR > 0.30, as recommended by PennCNV; a genome-wide LRR SD obtained from QuantiSNP greater than 3.50. To select CNVs with high confidence for downstream analyses, the following criteria were applied: (i) a maximum Bayes factor > 10 predicted by Quantisnp, (ii) called by both Quantisnp and PennCNV, and the breakpoints identified by the two algorithms should be within 2bp difference, (iii) with a physical length greater than 1kb and spanning three or more contiguous probes. For every gene/region that we were interested in a more profound quality control was performed. Samples with inconsistent CNV calls from two algorithms were further removed. The potential for population stratification was investigated by PCA undertaken with the EIGENSTRAT package [18]. Nine significant eigenvectors were identified based on the Tracy-Widom statistic (P<0.05), but none of them was significantly associated with case-control status (All P>0.05), suggesting that population stratification is not a confounder in our study.
Global burden analysis
We assessed the genome-wide CNV burden in patients and controls based on the number of CNVs (all CNVs, all genic CNVs, rare CNVs and rare genic CNVs) per genome. CNVs with a frequency ≤ 1.0% in our dataset were defined as rare CNVs. Genic CNVs were defined as CNVs overlapped with one or more genes (Genes were determined by RefSeq annotations (UCSC, Feb 2009, GRCh37/hg19) and gene boundaries were extended with a 10 kb flanking region on either side [19]). We first evaluated the burden of deletions and duplications together, followed by one or the other separately. Significant differences were determined by 1 × 106 permutations via PLINK [20].
Gene-based analysis
The numbers of patients and controls in whom a given gene was affected by CNVs were counted and compared. Permutation tests (1 × 106 iterations) were carried out for individual genes by PLINK and permutation correction for multiple testing for all genes was performed with the max (T) permutation (mperm) option of PLINK [20]. In addition, "OR", the full name of OR has mentioned above, 95 % confidence interval (CI) and P values for specific CNV-disrupted genes for the risk of cervical cancer were calculated by logistic regression in the allelic test using the SAS 9.3 software. Bayes false discovery probability (BFDP) calculation was performed to reduce the probability of false-positive findings from GWAS stage. Two levels of prior probability (0.05 and 0.005) was selected and prior OR of 1.5 was adopted, which was suggested by Wachoder et al. for a detrimental variant [21].BFDP <0.5 was used to indicate noteworthy findings.
Technical Replication
CNVs in CTDSPL and NEDD4L were technically validated using SybrGreen real-time quantitative PCR (RT-qPCR) on the ABI7900HT Fast Real-Time PCR System (Life Technologies, CA, USA). The Ct values were obtained with the SDS software v2.3 (Life Technologies). Copy numbers were calculated by the ΔΔCt method with normalization against both the reference gene and the samples without the corresponding CNV. In addition, ATPase 13A4 (ATP13A4) was excluded from the study as its CNV segment contained highly repeated sequences, which hindered successful design of efficient and specific primers. The following primers were used: CTDSPL (forward 5’-CTGGTGCTTTGAAGATACGG-3’, reverse 5’- AGCAATAGGCTTACAGAGGG-3’), NEDD4L (forward 5’-TGCTACTGACAGCCTAAATC-3’, reverse 5’-GGACCTCTGAGCCATAAAAG-3’) and RNase P (reference gene with two copies in diploid human genome, forward 5’- TATTCACAAAGAGCCCAGAG-3’, reverse 5’-GAAGGGTATGGGAAAACAAG-3’). The PCR reaction were performed in triplicates and comprised 10 ng of genomic DNA, 200 nM of each primer and PowerSYBR® Green PCR Master Mix (Cat. No. 4367659, Life Technologies). Samples were denatured at 95°C for 10 min followed by 40 cycles of 15 sec denaturation at 95°C and 1 min annealing at 60°C.
Independent Replication
CTDSPL and NEDD4L CNVs were independently determined in the replication cohort by using Custom Taqman Copy Number Assays (Life Technologies). Furthermore, CNVs of all the SybrGreen RT-qPCR validated samples were double validated and separately included as controls in the Taqman assays during each run. Data were obtained by the SDS software v2.3 (Life Technologies) and copy numbers were calculated using the Copy Caller v2.0 software with a calibrator sample without deletion of the CNV segments. The following oligonucleotides were used: CTDSPL forward primer 5’- GGTACAAATCTGATCCCGTCACT -3’, CTDSPL reverse primer 5’- CTTGCAAGCCATGGAGATGAG -3’, CTDSPL FAM-labeled probe 5’- CCCTTTTCCATAACATCAAATCC -3’, NEDD4L forward primer 5’- TTGGGTAAATCATGGCTTAAAACTCTCA -3’, NEDD4L reverse primer 5’- TCTGAATGCAGGGTGGGAAATAAAA-3’, NEDD4L FAM-labeled probe 5’- CTAGGTTCTTGCACATCTTTGC -3’. The RNase P primers and VIC-labelled probe included in the Taqman Copy Number Reference Assay (Cat. No.4403328, Life Technologies) were used as references and mixed with either the CTDSPL primer-probe mix or the NEDD4L primer-probe mix. Reactions were performed in duplicates using 10 ng of genomic DNA, target and reference gene primer-probe mix and Taqman Universal Master Mix (Cat. No. 4326614, Life Technologies) in a duplex format, by using the above-mentioned amplification cycles on the ABI7900HT Fast Real-Time PCR System.
Replication and combined statistical analysis
To account for the correlation between related subjects in the replication and combined datasets, the association analysis was performed using PedGenie that allows for a mixture of pedigree members and independent individuals to be analyzed together [18]. A Monte Carlo approach was employed to generate an empirical null distribution from which significance for a statistic of interest can be determined. Empirical 95 % CIs and P values were calculated using 1?1019 simulations.
Correlation between CNV and gene expression
Two hundreds and ninety three individuals with cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) from TCGA (http://www.cancergenome.nih.gov) with both CNV data and RNA-seq data available were included in this study. Level 3 CNV data detected from Affymetrix SNP 6.0 microarrays were downloaded. Segment mean values of the segments covering the 6.4kb deletion were extracted according to chromosome positions, and the copy number was calculated as (copy number value = 2*2^segment mean values). Copy number value between 1.7 and 2.3 was defined as no copy number variation, whereas <1.7 was defined as deletion and >2.3 was defined as duplication. In addition, RSEM- normalized results for RNA-seq data were downloaded. The association between the CNV of CTDSPL and expression of host gene and surrounding genes within 200kb was calculated by one-way ANOVA, Spearman correlation and linear regression analyses.
Cell culture
HEK293T was cultured in Dulbeccos modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco). Human epithelial cervical cancer cells (HeLa) purchased from America Type Culture Collection (ATCC) were cultured in 75 cm2 flasks in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich) supplemented with 10% (v/v) fetal bovine serum, 100 IU/mL penicillin and 100 mg/mL streptomycin. End1/E6E7 cells (human endocervical cells immortalized with human papillomavirus E6/E7) were obtained from ATCC and cultured in 75 cm2 flasks in keratinocyte serum-free medium supplemented with keratinocyte growth supplement, 100 U/ml penicillin and 100 mg/ml streptomycin. All cells were maintained at 37°C in a 5% CO2 incubator.
Lentiviral plasmid construction, lentiviral production and over-expression of ZNF263 cell line screening
The gene ZNF263 was PCR-amplified from a human cDNA library and was fused into a lentiviral vector containing an EF1α promoter, which co-expressed an EGFP fluorescence protein from a CMV promoter. The lentiviruses of ZNF263 were obtained by co-transfection with three viral packaging plasmids pLP1, pLP2 and pLP VSV-G into HEK293T using CaCl2 transfection. Virus was harvested 48 hr post-transfection. For viral transduction, cells were incubated with culture-medium- diluted viral supernatant for 24 hrs. At 72 hr following transduction, the EGFP-positive population was reached to more than 90 %.
Plasmid construction and luciferase assays in HEK293T stably overexpressing ZNF263
The CTDSPL intron 1-6367 bp fragment was PCR-amplified from 293T genomic DNA. To explore directionality of the regulatory element, we cloned the fragment upstream and downstream of SV40 of the pGL3-promoter vector (Promega), respectively. Inserts in each construct were verified by sequencing. Primer sequences are available on request. Constructs were transfected with equimolar amounts (800 ng) for luciferase reporter plasmids into HEK293T and HEK293-ZNF263 using lipofectamine 2000 reagent (Invitrogen), according to manufacturer’s instructions. Cells were harvested after 48h. Luminescence activity was measured with a Berthold lumat LB9507. Assays were performed in triplicate wells. Data represent at least three independent experiments. Statistical significance between experimental groups was assessed using a T test analysis. All analyses were performed using SAS 9.3, and a two tailed P value <0.05 was considered significant.
Plasmid construction and luciferase assays in HEK293T transiently overexpressing ZNF263
FMO3, GATA2 and IRF1 cDNA were synthesised by Sangon Biotech (Shanghai, China) and were cloned into pcDNA3.1 vectors which contain a CMV promoter, respectively. ZNF263 was PCR-amplified from a human cDNA library (Thermo Scientific CCSB-Broad Lentiviral Expression Library) and was fused into a pcDNA3.1 vector. The CTDSPL intron 1- 6367bp fragment was PCR-amplified from HEK293T genomic DNA, and then was inserted upstream of the promoter -luc+ transcriptional unit of pGL3-promoter vector (Promega). All constructs were verified by sequencing. Primer sequences are available on request. Constructs were co-transfected with equal weight amounts (500 ng) of luciferase reporter plasmids and equal weight amounts (500 ng) of cDNA over-expression plasmids into HEK293T cells using lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. Luciferase expression was normalized to 50 ng Renilla luciferase expression (pRL-SV40). Cells were harvested after 48h. Luminescence activity was measured with a Berthold Centro LB 960 Microplate Luminometer. Assays were performed in fourfold wells. Data represent at least three independent experiments. Statistical significance between experimental groups was assessed using T test. All analyses were performed using SAS 9.3, and a two tailed P value <0.05 was considered significant.
Plasmid construction and colony formation assay
CTDSPL cDNA fragment was amplified from Human Umbilical Vein Endothelial Cells (HUVEC) mRNA by RT-PCR. The product was extracted, purified and digested with Hind III and Xho I, and then was inserted to myc-tag fusion vector pcDNA3.1/myc-his-A. The base sequence of recombinant pcDNA3.1/myc-CTDSPL plasmid was in accordance with human CTDSPL cDNA fragment by agarose gel electrophoresis and DNA sequence analysis. CTDSPL shRNAs were purchased from Santa Cruz Biotechnology. All oligonucleotides and plasmids were transfected into cells using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer’s instructions.
Before genetic manipulation, cells were engineered to stably express firefly luciferase by transfection with pNifty-CMV- luciferase and selection with 500 μg/ml Zeocin. 1×103 cells were independently plated onto 60-mm tissue culture plates. After 10-14 days, visible colonies were fixed with 100 % methanol and stained with 0.1 % crystal violet in 20 % methanol for 15 min. Colony-forming efficiency was calculated as the number of colonies/plated cells × 100 %.
Immunohistochemistry
The immunohistochemistry assay was conducted on nude mouse xenograft tumour tissues to detect and score CTDSPL and Ki-67 expression using methods described previously [22]. CTDSPL and Ki-67 antibodies was obtained from Novus Biologicals. Statistical significance between experimental groups was assessed using a T-test. All analyses were performed using SAS 9.3, and a two tailed P value < 0.05 was considered significant.
Tumour xenograft models
All animal experiments were conducted with the approval of the Shanghai Jiao Tong University Institutional Committee for Animal Research and in conformity with national guidelines for the care and use of laboratory animals. Cells infected with a luciferase-encoding lentivirus (1×106) were inoculated subcutaneously into nude mice. There were 8 mice in each experimental group. Tumour volume (V) was monitored by measuring tumour length (L) and width (W) with callipers and then calculated with the formula (L×W2) × 0.5. Statistical significance between experimental groups was assessed using a T- test. All analyses were performed using SAS 9.3, and a two tailed P value <0.05 was considered significant.
Ctdspl-knockout Mice
The Ctdspl knockout (KO) mice (Ctdspl+/-) on C57BL/6 genetic background were generated by using the CRISPR/Cas9 system at Cyagen Biosciences Inc. (Shanghai, China). The mouse Ctdspl (GenBank accession number: NM_133710.3; Ensembl: ENSMUSG00000047409) is located on mouse chromosome 9. Exon 2 was selected as target site. Cas9 mRNA and gRNA generated by in vitro transcription were then injected into fertilized eggs for KO mouse productions. The founders were genotyped by PCR followed by DNA sequencing analysis. The genotypes of the Ctdspl+/- mice were confirmed by examining the PCR products using the following primers: Forward 5′- GATGCCTCAGCTTTGTCCTTGG-3′ and Reverse 5′-TTCTACCCTGTGGATTCTGAGGCTTG-3′. The positive founders were breeding to the next generation which was genotyped by PCR and DNA sequencing analysis. All the mice were maintained under specific-pathogen-free conditions. All the animal experiments were approved by the Institutional Animal care and Use Committee (IACUC) of Shanghai Jiao Tong University, School of Medicine.
Results
Association between CNVs and cervical cancer risk
Principal component analysis (PCA) using 17,386 markers in low linkage disequilibrium (LD) (pair-wise r2<0.02) showed that there was no statistically significant difference between cases and controls (P = 0.330) (Figure 1), suggesting no batch effect. Meanwhile, the quantile-quantile plot shows minimal evidence of genomic inflation (λ = 1.031), suggesting no systematic bias [2]. To maximize the finding of potential cervical cancer-associated CNVs, the two algorithms-QuantiSNP 2.0 [16] and PennCNV [15] were used for identifying CNVs from the signal intensity data of the SNP microarray (log-R ratio and B-allele frequency). After initial quality control (Methods), 973 cases with 11,056 autosomal CNVs and 3,485 controls with 33,492 autosomal CNVs were included for global burden analysis in the discovery stage. In order to improve prediction accuracy, as to every specific gene/region that we were interested, samples with inconsistent CNV calls from two algorithms were further removed. Therefore, the number of samples included in the final analysis varied across specific genes/regions.
Cervical cancer patients had a higher total genome-wide burden of CNVs (all categories), all genic CNVs, rare CNVs and rare genic CNVs than controls (fold change: 1.18, 1.16, 1.19, 1.09 respectively, all P< 0.05, (Table 1). The difference between patients and controls was particularly strong for duplications (fold change: 1.24, 1.22, 1.25, 1.19 for all duplications, all genic duplications, rare duplications and rare genic duplications respectively, all with a P < 0.05, (Table 1). All analyses, except for rare genic deletions (fold change = 1.05, P = 0.13), reached statistical significance (P < 0.05).
Table 1: Frequency of CNVs in cases versus controls
|
Type |
Category |
Cases (%) |
Control (%) |
Fold change |
P value |
|
Total |
All |
11.36 |
9.64 |
1.18 |
<1.0×10-6 |
|
All genic |
4.28 |
3.69 |
1.16 |
<1.0×10-6 |
|
|
Rare |
3.29 |
2.76 |
1.19 |
<1.0×10-6 |
|
|
Rare genic |
1.45 |
1.33 |
1.09 |
4.4×10-3 |
|
|
Deletions |
All |
9.15 |
7.85 |
1.17 |
<1.0×10-6 |
|
All genic |
3.13 |
2.74 |
1.14 |
<1.0×10-6 |
|
|
Rare |
2.35 |
2 |
1.17 |
<1.0×10-6 |
|
|
Rare genic |
0.97 |
0.92 |
1.05 |
0.13 |
|
|
Duplications |
All |
2.21 |
1.79 |
1.24 |
<1.0×10-6 |
|
All genic |
1.15 |
0.95 |
1.22 |
<1.0×10-6 |
|
|
Rare |
0.94 |
0.75 |
1.25 |
<1.0×10-6 |
|
|
Rare genic |
0.48 |
0.41 |
1.19 |
8.0×10-4 |
*P values were inferred using 1 × 106 permutations by PLINK
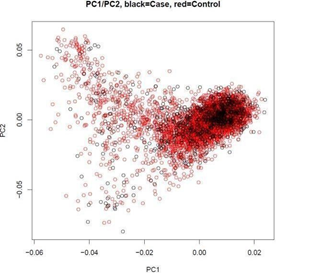
Figure 1: Principal component (PC)1 plotted against PC2 generated by principal component analysis (PCA). The black circles represent cervical cancer patients and the red circles represent controls
We then explored whether individual genes impacted by deletions or duplications were associated with susceptibility to cervical cancer. The strongest association was found for a 6367bp deletion in intron 1 of the CTD small phosphatase like gene (CTDSPL) (chr3: 37979882- 37986249), which was found in 7.4 % of cervical cancer cases and 2.6 % of controls (odds ratio OR = 2.61, 95% confidence interval CI = 1.91-3.56, P = 1.5×10-9, BFDP <0.5 when the prior probability is 0.5 or 0.05) (Table 2). Associations with deletions in ATP13A4 (OR = 2.31, 95% CI = 1.55-3.45, P = 4.3×10-5) and NEDD4L (OR
= 4.09, 95% CI = 2.07-8.05, P = 4.6×10-5) also reached statistical significance, but failed correction for multiple testing when the prior probability is 0.05 and 0.5, respectively (Table 2). To verify the accuracy of CNV calls, we reanalyzed a proportion of samples using both real-time quantitative PCR (RT-qPCR) and Custom Taqman Copy Number Assays (Life Technologies). Deletion of ATP13A4 was excluded from further investigation as it was unable to be designed for RT-qPCR or Taqman assay. Both the SybrGreen RT-qPCR and Custom Taqman Assays showed 100% concordance with the CNV calls from GWAS data on 99 cervical cancer cases tested for CTDSPL and 75 cervical cancer cases tested for NEDD4L based on the SNP arrays.
Table 2: CNV-disrupted genes associated with cervical cancer identified in the discovery phase
|
Frequency |
Association‡ |
BFDP§ |
||||||||
|
CNV |
Position* |
Type |
Case |
Control |
EMP† |
OR |
95%CI |
P |
0.5 |
0.05 |
|
CTDSPL |
chr3: 37979882- 37986249 |
deletion |
0.07 |
0.03 |
<1.0×10-6 |
2.6 1 |
1.91-3.56 |
1.5×10-9 |
3.0×10-4 |
0.03 |
|
ATP13A4 |
chr3:193119865 -193272696 |
deletion |
0.04 |
0.02 |
1.0×10-5 |
2.3 1 |
1.55-3.45 |
4.3×10-5 |
0.28 |
0.98 |
|
NEDD4L |
chr18:55711779 |
deletion |
0.02 |
0.005 |
7.0×10-5 |
4.0 |
2.07-8.05 |
4.6×10-5 |
0.71 |
1.00 |
|
-56065389 |
9 |
|||||||||
OR, Odds ratio; CI, confidence interval; BFDP, Bayes false discovery probability.
* Genomic positions were based on human genome assembly 19 (hg19).
† EMP represented P values calculated by PLINK using permutation tests (1,000,000 iterations).
‡ Odds ratio, 95% confidence interval and P values for the deletions were calculated by logistic regression in the allelic test using SAS.
Calculated as described previously [21].
The association with deletion in CTDSPL was further replicated in an independent study of 1,396 cervical cancer subjects and 1,057 controls (OR = 1.31, 95% CI = 1.05-1.65, P = 0.017) from the Swedish population (Methods, Table 3). The frequency of the deletion was 0.17 and 0.14 in the cancer patients and controls, respectively. In contrast, no association was observed between deletion in NEDD4L and cervical cancer risk in the replication series (OR = 1.04, 95% CI = 0.48–2.26, P= 0.92). Using the combined discovery and replication data, the deletion in CTDSPL showed an OR = 2.54 (95% CI = 2.08- 3.12, P = 2.0×10-19) with risk of cervical cancer. No heterogeneity for the association of CTDSPL deletion with cervical cancer risk was noted by tumour stage (CIS vs invasive cancer) (P = 0.12).
Table 3: Association results of CTDSPL-6367bp deletion with cervical cancer risk in the combined analysis
|
Number (Frequency) |
Association* |
||||
|
Case |
Control |
OR |
95%CI |
P |
|
|
overall By Genotype |
2290(0.13) |
4245(0.05) |
2.54 |
2.08-3.12 |
2.0×10-19 |
|
0 copy of del |
1990(0.87) |
4016(0.95) |
Ref |
Ref |
|
|
1 copies of del |
272(0.12) |
208(0.05) |
2.64 |
2.14-3.26 |
1.6×10-19 |
|
2 copies of del |
28(0.01) |
21(0.005) |
2.69 |
0.81-8.97 |
0.1 |
|
By study |
|||||
|
Discovery |
894(0.07) |
3188(0.03) |
2.61 |
1.91-3.56 |
1.5×10-9 |
|
Replication |
1396(0.17) |
1057(0.14) |
1.31 |
1.05-1.65 |
0.02 |
|
By tumour stage |
|||||
|
Carcinoma in situ |
2096(0.13) |
4245(0.05) |
2.45 |
1.99-3.02 |
3.7×10-17 |
|
Invasive cancer |
194(0.19) |
4245(0.05) |
3.58 |
2.32-5.53 |
8.6×10-9 |
del, deletion; OR, odds ratio; CI, confidence interval
* Odds ratio, 95% confidence interval and P values for the deletions were calculated by Pedgenie which corrected for relatedness except for the discovery phase in which odds ratio, 95% confidence interval and P values were calculated by logistic regression in the allelic test as there were no related subjects.
The effect of CTDSPL deletion on transcription
RNA-seq data in cervical cancer tissues from 293 individuals in The Cancer Genome Atlas (TCGA) (http://www.cancergenome.nih.gov) indicated that the 6367bp deletion was significantly associated with decreased expression of CTDSPL (Spearman coefficient = 0.54, p = 1.4×10-23), supporting that the deletion affects the transcription of CTDSPL (Methods). There are 3 other genes (villin like [VILL], phospholipase C delta 1[PLCD1] and deleted in lung and esophageal cancer 1 [DLEC1]) within the 200kb range of the deletion. We analyzed the data from TCGA to examine the association between the CTDSPL CNV and the expression level of these candidate genes (Methods). One-way ANOVA results suggested that expression level of CTDSPL showed more significant association with CNV of CTDSPL than the other genes (7.3×10-19 vs 0.02, 0.001 and 0.75, respectively) (Figure 2, Table 4). In addition, the significance of the correlation with VILL and PLCD1 did not remain in the linear regression analysis, while CTDSPL itself still showed remarkable significance (5.1×10-9, Table 5). Therefore, we speculated that the deletion of CTDSPL may be more likely to affect the transcription of CTDSPL itself rather than the surrounding genes.
Table 4: The association between the deletions that cover the 6367bp fragment in CTDSPL and expression levels of surrounding genes
|
Gene |
Deletion (N=91) |
Normal (N=187) |
Duplication (N=15) |
Pa |
|
CTDSPL |
840.7 ± 379.8 |
1380.2 ± 555.8 |
1712.3 ± 653.0 |
7.3×10-19 |
|
VILL |
438.6 ± 619.7 |
713.9 ± 884.0 |
683.4 ± 705.2 |
0.02 |
|
PLCD1 |
396.3 ± 302.1 |
512.9 ± 523.7 |
861.4 ± 868.1 |
0.001 |
|
DLEC1 |
40.4 ± 227.3 |
28.2 ± 105.0 |
15.5 ± 34.0 |
0.75 |
a Calculated by one-way ANOVA.
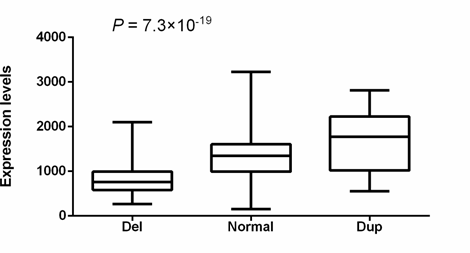
Figure 2: Correlation between CNV and CTDSPL transcriptional level. 293 cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) from The Cancer Genome Atlas (TCGA) (http://www.cancergenome.nih.gov) with both copy number variation data and RNA-seq data available were included. Statistical significance between groups was assessed using one-way ANOVA.
Table 5: Linear regression analysis between the deletions that cover the 6367bp fragment in CTDSPL and expression levels of surrounding genes
|
Gene |
Betaa |
Pa |
|
CTDSPL |
0.00028 |
5.1×10-9 |
|
VILL |
0.00002 |
0.55 |
|
PLCD1 |
0.00009 |
0.1 |
|
DLEC1 |
-0.00012 |
0.47 |
aCalculated by linear regression analysis.
As shown in (Figure 3), according to the Encyclopedia of DNA Elements (ENCODE) data [23], 6 transcriptional factors were predicted to bind to the CTDSPL intron 1-6367bp fragment (chr3:37979882-37986249), i.e. upstream transcription factor 2 (USF2) (chr3:37,981,985-37,982,248), activating transcription factor 3 (ATF3) chr3:37,982,058-37,982,290), upstream transcription factor 1 (USF1) (chr3:37,982,074-37,982,317), zinc finger protein 263 (ZNF263) (chr3:37,982,058- 37,982,290), binding protein 2 (GATA2) (chr3:37,983,211-37,983,617) and interferon regulatory factor 1(IRF1) (chr3:37,986,122- 37,986,432). All of them are expressed in cervical cells [24]. Three DNase-sensitive sites were found to be overlapping with the binding sites of the above five transcriptional factors, respectively, which are regulatory regions in general and promoters in particular [23]. ZNF263, GATA2 and IRF1 have been implicated in cancer development [24-39]. The regulatory role of this fragment was examined by luciferase reporter assays using a construct containing the fragment upstream of the SV40 promoter (Supplementary Methods). Relative promoter activity was determined in HEK293T cells that overexpress ZNF263, GATA2 and IRF1, respectively. Flavin containing monooxygenase 3 (FMO3), a transcription factor predicted to have no binding site in intron 1 of CTDSPL and is expressed in cervical cells [24], was set as an additional control. The location of the CTDSPL-6367bp fragment relative to the SV40 promoter (upstream or downstream) did not affect the transcriptional activity (Supplementary Figure 1). The construct containing the CTDSPL-6367bp fragment upstream of the SV40 promotor generated higher luciferase signals as compared to the PGL3-Control without the CTDSPL- 6367bp fragment, when co-transfected with ZNF263, GATA2 or IRF1 cDNA to HEK293T cells (P = 0.006, P = 0.035, P = 0.016, respectively) (Figure 4). However, there was no significant difference in the luciferase signals between the construct containing the CTDSPL-6367bp fragment and PGL3-Control when co-transfected with FMO3 cDNA to HEK293T (P > 0.05). These results indicate that the enhancer activity of the CTDSPL intron 1-6367bp fragment depends on the expression of ZNF263, GATA2 or IRF1.
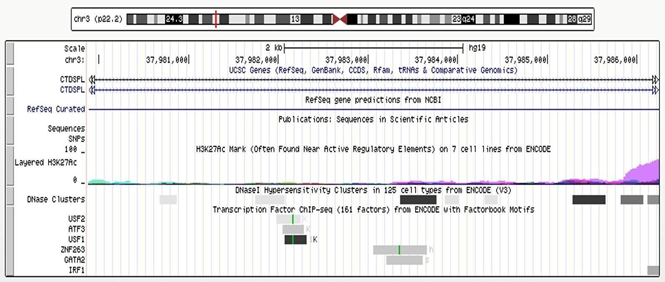
Figure 3: Genomic region at chr3:37979382-37986749 The track of “layered H3K27Ac” shows the levels of enrichment of the H3K27Ac histone mark across the genome as determined by a ChIP-seq assay. The H3K27Ac histone mark is the acetylation of lysine 27 of the H3 histone protein, and it is thought to enhance transcription possibly by blocking the spread of the repressive histone mark H3K27Me3. The track of “DNase clusters” shows DNase hypersensitive areas assayed in a large collection of cell types by the ENCODE project.
Regulatory regions in general, and promoters in particular, tend to be DNase-sensitive. The track at the bottom shows regions of transcription factor binding derived from a large collection of ChIP-seq experiments performed by the ENCODE project, together with DNA binding motifs identified within these regions by the ENCODE Factorbook repository [23].
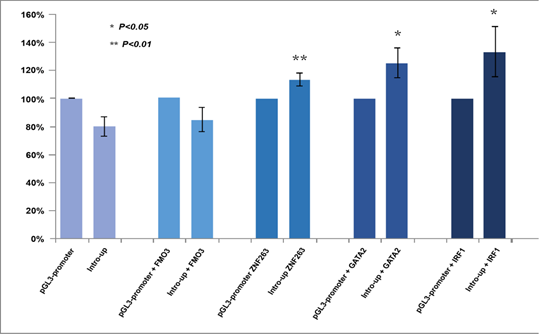
Figure 4: Luciferase signals of two pGL3 constructs in HEK293T cells which overexpressed FMO3, ZNF263, GATA2 and IRF1, respectively. Statistical significance between experimental groups was assessed using a T-test.
Function of CTDSPL in cervical cancer
To determine the function of CTDSPL in human cervical cancers, we ectopically expressed CTDSPL in HeLa cells lacking CTDSPL (Methods). HeLa cells expressing CTDSPL showed a significant decrease in colony-forming ability compared with cells expressing vector control (Figure 5A, B). Next, we examined the role of CTDSPL on tumour growth (Methods). HeLa cells expressing CTDSPL, or vector control, were injected subcutaneously into immunocompromised mice. Compared with control groups, mice injected with HeLa cells expressing CTDSPL exhibited a significant reduction in tumour volume (Figure 5C-E). Histologic analysis confirmed that all mice bearing tumours expressing CTDSPL showed a significant down- regulation of Ki-67 (Figure 5F). Moreover, we used two distinct short hairpin RNAs (shRNA1 and shRNA2; hereafter referred to collectively as shCTDSPL) to ablate CTDSPL expression in luciferase-expressing End1/E6E7 cells (Figure 5G). We then examined the effect of CTDSPL depletion on tumourigenicity by subcutaneous injection into mice with End1/E6E7 cells transduced with shCtrl or shCTDSPL (Methods). We observed that the mice implanted with control cells were viable and did not develop tumours when sacrificed, whereas CTDSPL-depleted immortalized End1/E6E7 could form tumours in NOD-SCID mice (Figure 5H). The tumor burden in the whole cohort is shown in Figure 5I.
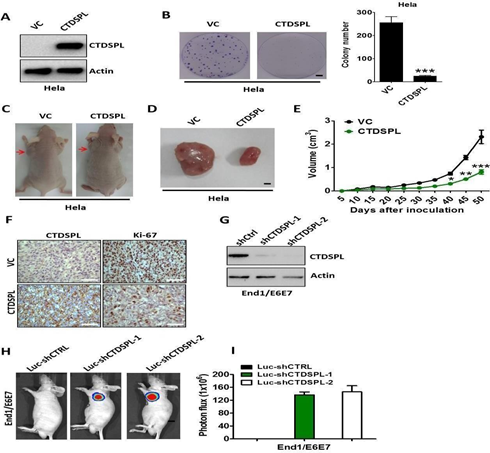
Figure 5: (A) Western blot analysis of CTDSPL expression in Hela cells transfected with pcDNA3.1-CTDSPL or vector control, respectively. Actin served as the loading control. (B). Colony formation ability of Hela cells after transfection of pcDNA3.1-CTDSPL or vector control. Data are means of three independent experiments ± SEM. ***, P < 0.001. (C) and (D). Representative images of xenograftsand tumours originated from Hela CTDSPL-overexpressing or vector control cells on the 50 days are shown. (E). Growth curve of CTDSPL-overexpressing or vector control cells-derived subcutaneous tumour xenografts. *, P < 0.05; **, P < 0.01; ***, P <0.001. (F).
The level of CTDSPL and Ki-67 was examined by IHC analysis in CTDSPL-overexpressing or vector control cells-derived tumour xenografts. Representative images of IHC are shown. (G). Western blot analysis of CTDSPL expression in End1/E6E7 cells transfected with two independent luciferase-encoding CTDSPL shRNA or control shRNA. Actin served as the loading control. (H). Representative pseudocolour bioluminescence images of mice bearing CTDSPL-depleted End1/E6E7 cells or control cells. (I). The tumor burden in the whole cohort of 8 mice/group. Statistical significance between experimental groups was assessed using a T-test.
Ctdspl-/- mice
The genotypes of the Ctdspl+/- mice were confirmed by examining the PCR fragments. A proximate 860 base pair PCR fragment was generated by amplifying the genomic DNA from wildtype mice. The mutant mice carried a 171-base-pair deletion in Ctdspl, thus the fragment was about 690 base pairs (Supplementary Figure 2). The F0 generation were mated to obtain F1 generation mice, homozygous for the deletion (Supplementary Figure 3). Upon examination one year later, the mice showed no sign of tumour.
Discussion
We have performed the first genome-wide CNV study of cervical cancer. We found that a 6367bp deletion in intron 1 of CTDSPL was associated with 2.54-fold increased risk of cervical cancer. This fragment is likely to enhance CTDSPL transcription with presence of transcriptional factor ZNF263, GATA2 or IRF1. In vitro and in vivo studies indicate that CTDSPL is a new tumour suppressor gene for cervical cancer. No frequency data can be found for the detected 6367bp deletion in intron 1 of CTDSPL in the 1000 Genome Project [40]. However, the CNV esv2676043 (chr3:37978417- 37986927) identified in the 1000 Genome Project which covers the CTDSPL-6367bp deletion had a frequency of 0.02, 0.06, 0.09 and 0.05 in the African population, American population, European population and Asian population, respectively. CTDSPL encodes a protein phosphatase that dephosphorylates RB1, halting the cell cycle at the G1/S boundary, thereby controlling cell proliferation [22]. It is conserved from yeast to human. CTDSPL is frequently deleted in cervical tumour and cervical intraepithelial neoplasia (CIN). The deletions in CTDSPL have been found to be significantly higher in squamous cell carcinoma (SCC) with metastases than in SCC without metastases, and decreased expression was more frequent in SCC with metastases as compared to SCC without metastases [41]. Reduced expression of CTDSPL in SiHa and CaSki along with tumour suppressive ability has been reported in both in vitro and in vivo systems [42]. The promoter of CTDSPL is highly methylated in cervical cancer [43]. Furthermore, CTDSPL deletion is associated with poor prognosis of cervical cancer patients [43].
In addition, we constructed Ctdspl gene knockout mice. The F0 generation were mated to obtain F1 generation mice, homozygous for the deletion. Upon examination one year later, the mice showed no sign of tumour. This indicates that knocking out the Ctdspl gene alone will not lead to cervical cancer. Instead, HPV might need to first trigger the initiation of tumour development, and the Ctdspl gene deletion will then increase the rate of tumour development. However, this hypothesis needs to be verified by analysis of Ctdspl-/- gene knockout mice carrying high-risk HPV infection.
Up to now, little is known about the function of ZNF263, except that it was predicted to have a repressive effect on gene transcription and often binds intragenic regions [25]. Our finding that ZNF263 upregulated CTDSPL provides new clues to ZNF263 function. Chen et al. reported that ZNF263 was upregulated in the blood of hepatocellular carcinoma (HCC) patients compared with the healthy volunteers [26], which implies that ZNF263 may play a role in the pathogenesis of HCC.
GATA2, a member of GATA family of transcription factors, is expressed principally in hematopoietic cell lineages, with a particularly prominent expression in early progenitors, as well as in megakaryocytes and in mast cell lineages [27]. Mutations in this gene have been related to various hematological malignancies, particularly in MDS/AML (myelodysplastic syndrome/acute myeloid leukemia) familial aggregations [28]. Recently, the functions of GATA2 as an oncogene in different types of human cancer have also been reported [29,44]. For example, GATA2 silencing could decrease cell migration and tissue invasion in prostate cancer [29]. Willman et al. presented convincing evidence that IRF1 was tumor suppressor gene [35]. Loss of heterozygosity (LOH) at the IRF1 locus occurs frequently in human gastric cancer [36]. In addition, Harada et al. observed alternative splicing of IRF1 mRNA, producing nonfunctional IRF1 protein at high frequencies in patients with myelodysplastic syndrome and acute myelogenous leukemia [38]. Several limitations to our study should be recognised. First, the technical validation of CNVs in the present study was limited. Ideally, a random sample of both cases and controls should have been taken for technical validation, oversampling those with the deletion. Unfortunately, this was not possible due to limited access to the DNA samples of the controls. For the technical validation we therefore oversampled subjects carrying the deletion and selected subjects at random without the deletion. Second, there are no SNPs on the Illumina HumanOmniExpress BeadChip that cover DEFB4 in our study. Therefore, we could not evaluate the association between DEFB4 copy number and susceptibility to cervical cancer.
In summary, we found that a 6367bp deletion in intron 1 of CTDSPL was associated with 2.54-fold increased risk of cervical cancer. This CNV is one of the strongest genetic risk variants identified so far for cervical cancer. This deletion removes the binding sites of ZNF263, GATA2 and IRF1, and hence downregulates the transcription of CTDSPL. In vitro and in vivo studies suggest that CTDSPL is a tumour suppressor gene for cervical cancer.
Conclusions
These findings indicate that CTDSPL is a tumour suppressor gene for cervical cancer and the 6367bp deletion downregulates
CTDSPL transcription by removing binding sites of ZNF263, GATA2 and IRF1.
Declarations
Ethics approval and consent to participate
The original collection of samples and study was approved by the Uppsala Ethics Committee with ref no Ups Dnr 97085. Informed consent was obtained from all subjects included in the study.
Consent for publication
Participants signed informed consent regarding publishing their data.
Data availability statement
The data that supports the findings of this study are available in the supplementary material of this article. The raw data cannot be made publicly available due to privacy or ethical restrictions.
Conflict of interest
The authors declare no competing interests.
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSFC) grants (81672563 DC, 81773027 DZ), the Youth Eastern Scholar (QD 2015006 DC), The Swedish Cancer Society (UG), the Medical Faculty of Uppsala University (DC) and the Swedish Research Council (UG). Illumina genotyping was performed by the SNP&SEQ
Technology Platform in Uppsala. The platform is part of Science for Life Laboratory at Uppsala University and supported as a national infrastructure by the Swedish Research Council.
Acknowledgements
The authors thank all of the participants in this research and support staff who made this study possible. We are grateful to all pathology clinics that enabled access to their archives.
Author’s contributions
DC designed the study, performed overall project management and drafted the initial manuscript; DC, HW and GU supervised the research. GU, DC and DZ obtained financial support. ZL and DZ performed the CNV analysis and had lots of inputs in the modified manuscript; SW and HW performed the animal experiments; TC performed the SYBR and Taqman assays; YG performed the cell experiments; PM coordinated the TwinGene study; DZ, DC, GU, XR, YG, YD, JH, PZ, XZ, XC, JG, DX and HW contributed to the interpretation of the results. XC and WY helped the writing. All authors contributed to the final paper.
Materials & Correspondence
Correspondence and material requests should be addressed to
- Dan Department of Gynecology, Third Xiangya Hospital of Central South University, Changsha, China or Shanghai Fosun Pharmaceutical Industrial Development, Co., Ltd. Shanghai, China.
- Gyllensten Department of Immunology, Genetics and Pathology, Science for Life Laboratory Uppsala, Uppsala University, Uppsala, Sweden.
- Huibo Wang. Department of Neurosurgery, First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu,
References
- Castle PE, Einstein MH, Sahasrabuddhe Cervical cancer prevention and control in women living with human immunodeficiency virus. CA Cancer J Clin 71 (2021): 505-526.
- Chen D, Juko-Pecirep I, Hammer J, et al. Genome-wide association study of susceptibility loci for cervical J Natl Cancer Inst 105 (2013): 624-633.
- Shi Y, Li L, Hu Z, et A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet 45 (2013):918-922.
- Sebat J, Lakshmi B, Troge J, et Large-scale copy number polymorphism in the human genome. Science 305 (2004): 525-528.
- Iafrate AJ, Feuk L, Rivera MN, et Detection of large-scale variation in the human genome. Nat Genet 36 (2004): 949- 951.
- Eichler EE, Flint J, Gibson G, et Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11 (2010): 446-450.
- Zhou Y, Hao Y, Li Y, et Amplification and up-regulation of MIR30D was associated with disease progression of cervical squamous cell carcinomas. BMC Cancer 17 (2017): 230.
- Lee M, Nam ES, Jung SH, et 1p36.22 region containing PGD gene is frequently gained in human cervical cancer. J Obstet Gynaecol Res 40 (2014): 545-553.
- Mendoza-Rodriguez M, Arreola H, Valdivia A, et al. Cellular retinol binding protein 1 could be a tumor suppressor gene in cervical Int J Clin Exp Pathol 6 (2013): 1817-1825.
- Ng G, Winder D, Muralidhar B, et al. Gain and overexpression of the oncostatin M receptor occur frequently in cervical squamous cell carcinoma and are associated with adverse clinical J Pathol 212 (2007): 325-334.
- Lee BH, Roh S, Kim YI, et al. Difference of Genome-Wide Copy Number Alterations between High-Grade Squamous Intraepithelial Lesions and Squamous Cell Carcinomas of the Uterine Cervix. Korean J Pathol 46 (2012): 123-130.
- Snijders PJ, van Duin M, Walboomers JM, et al. Telomerase activity exclusively in cervical carcinomas and a subset of cervical intraepithelial neoplasia grade III lesions: strong association with elevated messenger RNA levels of its catalytic subunit and high-risk human papillomavirus Cancer Res 58 (1998): 3812-3818.
- Mitra S, Mazumder Indra D, Basu PS, et al. Amplification of CyclinL1 in uterine cervical carcinoma has prognostic Mol Carcinog 49 (2010): 935-943.
- Abe S, Miura K, Kinoshita A, et al. Copy number variation of the antimicrobial-gene, defensin beta 4, is associated with susceptibility to cervical cancer. J Hum Genet 58 (2013):250-253.
- Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping Genome Res 17 (2007): 1665-1674.
- Colella S, Yau C, Taylor JM, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping Nucleic Acids Res 35 (2007): 2013-2025.
- Diskin SJ, Li M, Hou C, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping Nucleic Acids Res 36 (2008): 126.
- Allen-Brady K, Wong J, Camp NJ. PedGenie: an analysis approach for genetic association testing in extended pedigrees and genealogies of arbitrary BMC Bioinformatics 7 (2006): 209.
- Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum Nature 466 (2010): 368-372.
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage Am J Hum Genet 81 (2007): 559-575.
- Sholom Wacholder, Stephen Chanock, Montserrat Garcia-Closas, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology J Natl Cancer Inst 96 (2004): 434-442.
- Luo H, Chen Z, Wang S, et al. c-Myc-miR-29c-REV3L signalling pathway drives the acquisition of temozolomide resistance in Brain 138 (2015): 3654-3672.
- Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 489 (2012):57-74.
- Klijn C, Durinck S, Stawiski EW, et A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 33 (2015): 306-312.
- Frietze S, Lan X, Jin VX, et Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J Biol Chem 285 (2010): 1393-1403.
- Chen J, Qian Z, Li F, et Integrative Analysis of Microarray Data to Reveal Regulation Patterns in the Pathogenesis of Hepatocellular Carcinoma. Gut Liver 11 (2017): 112-120.
- Vicente C, Conchillo A, Garcia-Sanchez MA, et al. The role of the GATA2 transcription factor in normal and malignant Crit Rev Oncol Hematol 82 (2012):1-17.
- Kazenwadel J, Secker GA, Liu YJ, et Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 119(2012): 1283-1291.
- Chiang YT, Wang K, Fazli L, et al. GATA2 as a potential metastasis-driving gene in prostate cancer. Oncotarget 5 (2014): 451-461.
- He B, Lanz RB, Fiskus W, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor Proc Natl Acad Sci U S A 111 (2014): 18261-18262.
- Bohm M, Locke WJ, Sutherland RL, et al. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated Oncogene 28 (2009):3847-3856.
- Wang Y, He X, Ngeow J, et al. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum Mol Genet 21 (2012): 569-
- Kumar MS, Hancock DC, Molina-Arcas M, et al. The GATA2 transcriptional network is requisite for RAS oncogene- driven non-small cell lung cancer. Cell 149 (2012): 642-655.
- Wang Z, Yuan H, Sun C, et al. GATA2 promotes glioma progression through EGFR/ERK/Elk-1 pathway. Med Oncol 32 (2015):
- Willman CL, Sever CE, Pallavicini MG, et al. Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemic Science 259 (1993):968-971.
- Tamura G, Sakata K, Nishizuka S, et Allelotype of adenoma and differentiated adenocarcinoma of the stomach. J Pathol 180 (1996): 371-377.
- Nozawa H, Oda E, Ueda S, et al. Functionally inactivating point mutation in the tumor-suppressor IRF-1 gene identified in human gastric cancer. Int J Cancer 77 (1998): 522-527.
- Harada H, Kondo T, Ogawa S, et Accelerated exon skipping of IRF-1 mRNA in human myelodysplasia/leukemia; a possible mechanism of tumor suppressor inactivation. Oncogene 9 (1994): 3313-3320.
- Nozawa H, Oda E, Nakao K, et Loss of transcription factor IRF-1 affects tumor susceptibility in mice carrying the Ha-ras transgene or nullizygosity for p53. Genes Dev 13 (1999): 1240-1245.
- Genomes Project C, Abecasis GR, Altshuler D, et A map of human genome variation from population-scale sequencing. Nature 2010;467(7319):1061-1073.
- Kashuba VI, Pavlova TV, Grigorieva EV, et High mutability of the tumor suppressor genes RASSF1 and RBSP3 (CTDSPL) in cancer. PLoS One 4 (2009): 5231.
- Anedchenko EA, Kiseleva NP, Dmitriev AA, et al. [Tumor suppressor gene RBSP3 in cervical carcinoma: copy number and transcriptional level]. Mol Biol (Mosk) 41 (2007): 86-95.
- Mitra S, Mazumder Indra D, Bhattacharya N, et RBSP3 is frequently altered in premalignant cervical lesions: clinical and prognostic significance. Genes Chromosomes Cancer 49 (2010): 155-170.
- Gong C, Fan Y, Zhou X, et Comprehensive Analysis of Expression and Prognostic Value of GATAs in Lung Cancer. J Cancer 12 (2021): 3862-3876.
