Dynamic Regulation of MALAT1 in Leader Cell Formation and Collective Cancer Invasion
Article Information
Purnima Tyagi1, Akhilesh Kumar Saini1 & Jitendra Kumar1*
1Department of Molecular and Cellular Medicine, Institute of Liver and Biliary Sciences, New Delhi, India-110070.
*Corresponding Author: Jitendra Kumar, Research Associates-I, Department of Molecular and Cellular Medicine, Institute of Liver and Biliary Sciences, New Delhi, India-110070.
Received: 11 August 2023; Accepted: 25 September 2023; Published: 27 September 2023.
Citation: Purnima Tyagi, Akhilesh Kumar Saini & Jitendra Kumar. Dynamic Regulation of MALAT1 in Leader Cell Formation and Collective Cancer Invasion. Journal of Analytical Techniques and Research 5 (2023): 13-15.
Share at FacebookAbstract
The primary mechanism in cancer metastasis, collective invasion is becoming more widely recognized. Leader cells are specially trained cancer cells that invading the tumors. They are crucial in establishing invasion pathways, organizing follower cells, and facilitating the survival of cancer cells throughout the metastatic cascade. To aid group invasion, these leader cells activate a variety of mechanical, genomic, and metabolomic pathways. Leader cell development and function are influenced by stromal cells, matrix properties, genetic and epigenetic factors, and more.
Keywords
Leader Cells, Live Single Cell Analysis, Long Noncoding RNAs, MALAT1, FRET nano biosensor.
Leader Cells articles; Live Single Cell Analysis articles; Long Noncoding RNAs articles; MALAT1 articles; FRET nano biosensor articles.
Leader Cells articles Leader Cells Research articles Leader Cells review articles Leader Cells PubMed articles Leader Cells PubMed Central articles Leader Cells 2023 articles Leader Cells 2024 articles Leader Cells Scopus articles Leader Cells impact factor journals Leader Cells Scopus journals Leader Cells PubMed journals Leader Cells medical journals Leader Cells free journals Leader Cells best journals Leader Cells top journals Leader Cells free medical journals Leader Cells famous journals Leader Cells Google Scholar indexed journals Live Single Cell Analysis articles Live Single Cell Analysis Research articles Live Single Cell Analysis review articles Live Single Cell Analysis PubMed articles Live Single Cell Analysis PubMed Central articles Live Single Cell Analysis 2023 articles Live Single Cell Analysis 2024 articles Live Single Cell Analysis Scopus articles Live Single Cell Analysis impact factor journals Live Single Cell Analysis Scopus journals Live Single Cell Analysis PubMed journals Live Single Cell Analysis medical journals Live Single Cell Analysis free journals Live Single Cell Analysis best journals Live Single Cell Analysis top journals Live Single Cell Analysis free medical journals Live Single Cell Analysis famous journals Live Single Cell Analysis Google Scholar indexed journals Long Noncoding RNAs articles Long Noncoding RNAs Research articles Long Noncoding RNAs review articles Long Noncoding RNAs PubMed articles Long Noncoding RNAs PubMed Central articles Long Noncoding RNAs 2023 articles Long Noncoding RNAs 2024 articles Long Noncoding RNAs Scopus articles Long Noncoding RNAs impact factor journals Long Noncoding RNAs Scopus journals Long Noncoding RNAs PubMed journals Long Noncoding RNAs medical journals Long Noncoding RNAs free journals Long Noncoding RNAs best journals Long Noncoding RNAs top journals Long Noncoding RNAs free medical journals Long Noncoding RNAs famous journals Long Noncoding RNAs Google Scholar indexed journals MALAT1 articles MALAT1 Research articles MALAT1 review articles MALAT1 PubMed articles MALAT1 PubMed Central articles MALAT1 2023 articles MALAT1 2024 articles MALAT1 Scopus articles MALAT1 impact factor journals MALAT1 Scopus journals MALAT1 PubMed journals MALAT1 medical journals MALAT1 free journals MALAT1 best journals MALAT1 top journals MALAT1 free medical journals MALAT1 famous journals MALAT1 Google Scholar indexed journals FRET nano biosensor articles FRET nano biosensor Research articles FRET nano biosensor review articles FRET nano biosensor PubMed articles FRET nano biosensor PubMed Central articles FRET nano biosensor 2023 articles FRET nano biosensor 2024 articles FRET nano biosensor Scopus articles FRET nano biosensor impact factor journals FRET nano biosensor Scopus journals FRET nano biosensor PubMed journals FRET nano biosensor medical journals FRET nano biosensor free journals FRET nano biosensor best journals FRET nano biosensor top journals FRET nano biosensor free medical journals FRET nano biosensor famous journals FRET nano biosensor Google Scholar indexed journals Molecular articles Molecular Research articles Molecular review articles Molecular PubMed articles Molecular PubMed Central articles Molecular 2023 articles Molecular 2024 articles Molecular Scopus articles Molecular impact factor journals Molecular Scopus journals Molecular PubMed journals Molecular medical journals Molecular free journals Molecular best journals Molecular top journals Molecular free medical journals Molecular famous journals Molecular Google Scholar indexed journals Cellular Medicine articles Cellular Medicine Research articles Cellular Medicine review articles Cellular Medicine PubMed articles Cellular Medicine PubMed Central articles Cellular Medicine 2023 articles Cellular Medicine 2024 articles Cellular Medicine Scopus articles Cellular Medicine impact factor journals Cellular Medicine Scopus journals Cellular Medicine PubMed journals Cellular Medicine medical journals Cellular Medicine free journals Cellular Medicine best journals Cellular Medicine top journals Cellular Medicine free medical journals Cellular Medicine famous journals Cellular Medicine Google Scholar indexed journals metabolomic pathways articles metabolomic pathways Research articles metabolomic pathways review articles metabolomic pathways PubMed articles metabolomic pathways PubMed Central articles metabolomic pathways 2023 articles metabolomic pathways 2024 articles metabolomic pathways Scopus articles metabolomic pathways impact factor journals metabolomic pathways Scopus journals metabolomic pathways PubMed journals metabolomic pathways medical journals metabolomic pathways free journals metabolomic pathways best journals metabolomic pathways top journals metabolomic pathways free medical journals metabolomic pathways famous journals metabolomic pathways Google Scholar indexed journals metastatic cascade articles metastatic cascade Research articles metastatic cascade review articles metastatic cascade PubMed articles metastatic cascade PubMed Central articles metastatic cascade 2023 articles metastatic cascade 2024 articles metastatic cascade Scopus articles metastatic cascade impact factor journals metastatic cascade Scopus journals metastatic cascade PubMed journals metastatic cascade medical journals metastatic cascade free journals metastatic cascade best journals metastatic cascade top journals metastatic cascade free medical journals metastatic cascade famous journals metastatic cascade Google Scholar indexed journals long noncoding RNAs articles long noncoding RNAs Research articles long noncoding RNAs review articles long noncoding RNAs PubMed articles long noncoding RNAs PubMed Central articles long noncoding RNAs 2023 articles long noncoding RNAs 2024 articles long noncoding RNAs Scopus articles long noncoding RNAs impact factor journals long noncoding RNAs Scopus journals long noncoding RNAs PubMed journals long noncoding RNAs medical journals long noncoding RNAs free journals long noncoding RNAs best journals long noncoding RNAs top journals long noncoding RNAs free medical journals long noncoding RNAs famous journals long noncoding RNAs Google Scholar indexed journals
Article Details
Commentary
Collective invasion is increasingly becoming recognized as the primary mechanism in cancer spread. Leader cells are cancer cells that have been properly taught to infiltrate tumors. They are critical in building invasion paths, organizing follower cells, and allowing cancer cells to survive throughout the metastatic cascade. These leader cells use a range of mechanical, genetic, and metabolomic routes to aid in group invasion. Stromal cells, matrix characteristics, genetic and epigenetic variables, and other factors all influence leader cell formation and function. The development and metastasis of cancer have been shown to be significantly influenced by long noncoding RNAs (lncRNAs) [1]. Despite not having the ability to code for proteins, lncRNAs can regulate gene expression using a variety of methods. They play a role in post-transcriptional regulation of cytoplasmic RNA and post-translational regulation of chromatin-modifying enzymes, alternative splicing, and epigenetic regulation of chromatin- modifying enzymes. Numerous lncRNAs, such as urothelial cancer-associated 1 (UCA1) [2] and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) [3], have been found to control the spread of cancer. For instance, through controlling genes linked to metastasis and affecting splicing patterns, MALAT1 encourages metastasis in lung and other malignancies. By deactivating pro-metastatic transcription factors and controlling the epithelial-mesenchymal transition (EMT), MALAT1 has also been discovered to inhibit metastasis in breast and colon cancers. These studies highlight the complex roles that lncRNAs play in cancer and the difficulties in understanding how they work, calling for the development of new technologies to better comprehend how lncRNAs play important roles in cancer [4].
Investigating the function of MALAT1 in leader cell production and cancer invasion is the main goal of this study. The role of lncRNAs must be investigated using live single-cell biosensors with high spatio-temporal resolution because leader cells make up a very small portion of cancer cells. Miserably, current methods that lyse or fix samples, including RNA sequencing and RNA fluorescence in situ hybridization (FISH), are unable to shed light on the spatio-temporal dynamics of RNAs in cancer cells during collective cancer invasion. This study introduces a FRET nano-biosensor for dynamic analysis of lncRNAs in live single cells in both two-dimensional (2D) and three-dimensional (3D) invasion models to overcome this constraint. To improve the signal-to-noise ratio, this novel nano-biosensor combines the double-stranded lock nucleic acid (dsLNA) probe, the dual probe design, and FRET-based sensing. Utilising this FRET method, the nano-biosensor reduces background noise brought on by autofluorescence and thermal quencher probe dissociation, making it easier to detect endogenous RNA transcripts in living single cells. Transient knockdown experiments utilizing small interfering RNA (siRNA) and stimulation with transforming growth factor beta 1 (TGF-1) are used to validate the FRET signal of the biosensor. The nano-biosensor is effective in identifying lncRNA expressions in migrating cell monolayers, tumor organoids made from patient samples, and three-dimensional cancer spheroids. The study evaluates the expression of b-actin, MALAT1, and UCA1 transcripts in active bladder cancer cells using the FRET nano-biosensor. Fluorescent puncta in the FRET channel, which stand for one or more RNA transcripts, are used to detect the presence of these RNA transcripts. The distribution of RNA transcripts in live cells is also assessed by the investigation using nuclear and cytoplasmic markers. The co-localization of RNA transcripts with the nuclei is identified using confocal microscopy and a live cell nuclear dye. There were often more b-actin, MALAT1, and UCA1 transcripts in the cytoplasm than there were co-localized with the nucleus. Additionally, TGF-b therapy, a recognized EMT inducer, causes an overall rise in MALAT1 and UCA1 expression in the cells, especially in the nucleus. However, both the cytoplasmic and nuclear areas continue to express b-actin mRNA at the same levels.
The work uses a scratch cell migration assay, generally known as a wound healing assay, to examine the distribution of lncRNAs during collective cancer migration. This assay enables the study of lncRNA dynamics in a population of cancer cells travelling together. The cells in the study migrate coherently and organize into projecting points with a leader-follower structure, which is consistent with another research. After roughly eight hours of scratching, migration tips and leader cells start to form. Cells at the protruding tips were categorized in the study as leader cells, and cells behind them as follower cells. Surprisingly, leader cells were show to have considerably more MALAT1 expression than follower cells. The expressions of UCA1 and b- actin mRNA in leader and follower cells, however, do not alter noticeably.
Further in this study used siRNA therapy to reduced MALAT1 expression in order to investigate MALAT1 roles in collective cancer migration. The findings shows that transiently inhibited MALAT1 prevented the development of migration tips and messes up collective cell movement. The monolayer boundary slightly contracts, lowered the average migration rate. Furthermore, MALAT1 knockdown results in a non-migratory phenotype, cell morphology. In contrast, downregulating UCA1 does not result in a significant alteration of cell shape, and the rate of migration was equivalent to that of cells treated with control siRNA. Additionally, TGF-b treatment of moving monolayers causes the separation of cancer cells and breakdown of cell-cell adhesion. Notably, certain dissociated cells exhibit increased MALAT1 expression.
Through single-molecule tracking, the work explored MALAT1 in leader and follower cells further. It is possible to calculate the mean squared displacement (MSD) of molecules by monitoring the positions of transcripts over time. The cytoplasmic MALAT1 has a diffusivity of about 0.09 mm2/s, which is comparable to that of b-actin mRNA. It is interesting to note that the nuclei of leader cells exhibit a decreased diffusivity of MALAT1. MALAT1 in the cytoplasm of leader cells has a higher diffusivity (0.089±0.017 mm2/s), but MALAT1 in the cell nuclei has a lower diffusivity (0.050±0.011 mm2/s). The MALAT1 diffusivity in follower cells, however, is comparable in the cytoplasm (0.097±0.033 mm2/s) and nuclei (0.010±0.043 mm2/s). Furthermore, only around 50% of MALAT1 transcripts are mobile (diffusivity 0.005 mm2/s), yet this percentage is constant between leader and follower cells. Since leader cells have lesser diffusivity than follower cells, it is possible that MALAT1 in leader cells participated in different molecular complexes and activities. These results revealed that MALAT1 activation in leader cells varies during collective.
Conclusion
The significance of MALAT1 in leader cell formation and mass cancer invasion is highlighted by this study. The study sheds light on the dynamic control of lncRNAs in living single cells using a FRET nano-biosensor. The results showed that during collective cancer migration, MALAT1 expression is increased in leader cells as opposed to follower cells. The development of migration tips is disrupted and collective cell migration was impaired when MALAT1 is knock-down, highlighting the critical function of MALAT1 in orchestrating collective invasion. The study also shown that MALAT1 has a variable distribution and level of activity in leader cells, pointing to its participation in various molecular complexes and functions. Furthermore, MALAT1 expression is heterogeneous in bladder cancer samples and tumor organoids, indicating its relationship with invasive characteristics. Overall, this study reveals that the FRET nano-biosensor potential for study lncRNA dynamics and emphasizes the role of MALAT1 in bladder cancer invasion co-ordination. It is necessary to conduct more research to clarify the MALAT1 molecular mechanisms and roles, which could lead to the development of new cancer diagnostic and therapy.
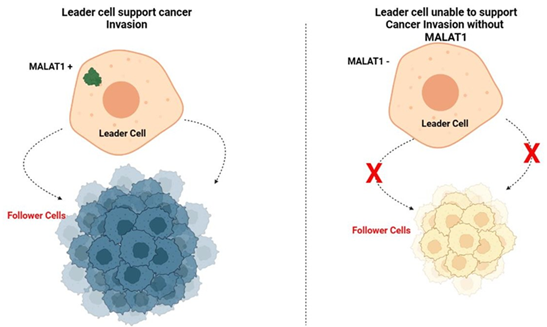
Figure 1: MALAT1 Expression in Leader Cells and its impact on Cancer Invasion.
Conflict of Interest
Authors declared no conflict of interest.
Fund
No funding support received.
References
- J Wang, J Zhang, H Zheng, et al. Neutral evolution of ‘non-coding’ complementary DNAs, Nature 431 (2004): 1-2.
- M Xue, W Chen, X Li. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer, J. Cancer Res. Clin. Oncol 142 (2016): 1407-1419.
- G Arun, S D Diermeier, D L Spector. Therapeutic Targeting of Long Non-Coding RNAs in Cancer, Trends Mol. Med 24 (2018): 257-277.
- N Zhu, M Ahmed, Y Li, et al. Long noncoding RNA MALAT1 is dynamically regulated in leader cells during collective cancer invasion, Proc. Natl. Acad. Sci 120 (2023): 2305410120.
