Denovo Structural Modeling and B cell and T cell Epitope Prediction against SARS-COV-2 PLpro to Cure COVID-19: Vaccinomics Based Approach
Article Information
Fabian JS van der Velden1,2, Priyen Shah3, Marie Voice4, Emma Lim1, Jethro Herberg3, Victoria Wright3, Tisham De3, Enitan D. Carrol5,6, Aakash Khanijau5, Andrew J Pollard7, Stéphane Paulus7, Ulrich von Both8, Laura Kolberg8, Clementien L Vermont9, Nienke N Hagedoorn9, Federico Martinón-Torres10,11,12, Irene Rivero Calle10, Philipp KA Agyeman13, Luregn J Schlapbach14, Maria Tsolia15, Irini Eleftheriou15, Marko Pokorn16, Mojca Kolnik16, Taco W Kuijpers17, Dace Zavadska18, Aleksandra Rudzate18, Nina A Schweintzger19, Werner Zenz19, Shunmay Yeung20, Michiel van der Flier21,22, Ronald de Groot21, Michael Levin3, Colin Fink4, Marieke Emonts1,2,23,*, on behalf of the PERFORM consortium**
1Great North Children’s Hospital, Paediatric Immunology, Infectious Diseases & Allergy, The Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle upon Tyne, United Kingdom
2Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, United Kingdom
3Wright-Fleming Institute, Section of Paediatric Infectious Disease, Imperial College London, London, United Kingdom
4Micropathology Ltd, University of Warwick, Warwick, United Kingdom
5Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, United Kingdom
6Alder Hey Children’s NHS Foundation Trust, Liverpool, United Kingdom
7Oxford Vaccine Group, Department of Paediatrics, University of Oxford, Oxford, United Kingdom
8Division Paediatric Infectious Diseases, Dr. Von Hauner Children’s Hospital, University Hospital, LMU, Munich, Germany
9Department of Pediatrics, Division of Pediatric Infectious Diseases & Immunology, Erasmus MC-Sophia Children’s Hospital, Rotterdam, The Netherlands
10Pediatrics Department, Translational Pediatrics and Infectious Diseases, Hospital Clínico Universitario de Santiago, Santiago de Compostela, Spain
11GENVIP Research Group, Instituto de Investigación Sanitaria de Santiago, Universidad de Santiago de Compostela, Santiago de Compostela, Spain
12Consorcio Centro de Investigacion Biomedicaen Red de Enfermedades Respiratorias (CIBERES), Madrid, Spain
13Department of Pediatrics, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland
14 Department of Intensive Care and Neonatology, and Children’s Research Center, University Children’s Hospital Zürich, Zürich, Switzerland
152nd Department of Pediatrics, National and Kapodistrian University of Athens, Children’s Hospital ‘P, and A. Kyriakou’, Athens, Greece
16University Children’s Hospital, University Medical Centre Ljubljana, Ljubljana, Slovenia
17Amsterdam University Medical Center, location Academic medical Center, Department of Pediatric Immunology, Rheumatology and Infectious Diseases, University of Amsterdam, Amsterdam, The Netherlands
18Department of Pediatrics, Rigas Stradina Universitate, Children’s Clinical University Hospital, Riga, Latvia
19Department of Pediatrics and Adolescent Medicine, Division of General Pediatrics, Medical University of Graz, Graz, Austria
20Clinical Research Department, Faculty of Infectious and Tropical Disease, London School of Hygiene and Tropical Medicine, London, United Kingdom
21Pediatric Infectious Diseases and Immunology, Amalia Children’s Hospital, Radboud University Medical Center, Nijmegen, The Netherlands
22Pediatric Infectious Diseases and Immunology, Wilhelmina Children’s Hospital, University Medical Center Utrecht, Utrecht, The Netherlands
23NIHR Newcastle Biomedical Research Centre, Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University, Newcastle upon Tyne, United Kingdom
*Corresponding author: Marieke Emonts, Great North Children’s Hospital, Paediatric Immunology, Infectious Diseases and Allergy, The Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle upon Tyne, United Kingdom.
**Members of the PERFORM Consortium are listed in the supplementary files
Received: 01 August 2023; Accepted: 09 August 2023; Published: 14 September 2023
Citation: Carlos Eliel Maya Ramirez, Sajida Ashraf, Faiza Irshad, Tabish Rehman, Moayad Shahwan, Muhammad Sufyan. Denovo Structural Modeling and B cell and T cell Epitope Prediction against SARS-COV-2 PLpro to Cure COVID-19: Vaccinomics Based Approach. Journal of Bioinformatics and Systems Biology. 6 (2023): 209-262.
Share at FacebookAbstract
Background: In December 2019, SARS COV-2 spread very rapidly and its outbreaks led to cause pneumonia and severe illness. For the first time, the virus was spotted in the city of China (Wuhan). Little is known about this virus, so the vaccine is needed to prevent COVID-19. Papain like protease (PLpro) is conserved among coronaviruses and have multifunctional activities like protease, deubiquitinase, deISGylating and interferon antagonism. Therefore, PLpro is a promising target for drug and vaccine development.
Methods: T cells (MHCI & MHCII) and B cell (Discontinuous & Linear) epitopes of SARS COV-2 PLpro were forecasted via computational and immunoinformatics approaches that show a crucial part in promoting immune responses against COVID-19. Online tools were used to analyze the allergenicity, physiochemical properties, antigenicity and structural details of PLpro.
Results: Fifty-three Linear B-cell epitopes were identified, of which ‘KPLEFGATSAALQP’ and ‘EDDYQGKPLEFGAT’ had higher score and antigenicity. Antigenic, non-allergenic and conserved (T cell) epitopes whichwere bounded to multiple alleles were selected. In total, twelve epitopes were taken (6 MHC I and 6 MHC II).From MHC class I ‘YHTTDPSFLGRY’ and MHC class II ‘PFVMMSAPPAQYELK’ had more antigenicity among T cell epitopes. Furthermore, the protection and stability of peptides were tested through digestion analysis.
Conclusion: Predicted epitopes could be served as vaccine candidate to eliminate COVID-19. Even so, the presented epitopes required to be validated experimentally to check its safety and immunogenic profile
Keywords
Papain like protease, Digestion analysis, Allergenicity, Antigenicity
Papain like protease articles; Digestion analysis articles; Allergenicity articles; Antigenicity articles
Article Details
1. Introduction
Coronaviruses are positive-sense RNA viruses which are widely spread among humans and many other species of mammals [1]. Two types of coronaviruses such as SARS-CoV [2-4] and MERS-CoV [5, 6] cause infection and more than 10,000 cases have been reported in the past two decades. In December 2019, numerous incidences of an unknown cause of COVID-19 occurred in Wuhan, Hubei, China, with the clinical presentations closely similar to viral pneumonia [7]. The cause of the cases of pneumonia was reported as a novel beta coronavirus, SARS-CoV- 2.The genome of SARS-CoV-2 includes 3-chymotrypsin like protease (3CLpro), papain-like protease (PLpro), RNA-dependant-RNA polymerase andstructural proteins (spike glycoprotein, Envelope, membrane and nucleoprotein).Sequence analysis of SARS-CoV-2 revealed, receptor- binding domain(RBD) and receptor binding motif(RBM) together interact with the receptor angiotensin enzyme 2(ACE2)[8, 9].
SARS-CoV-2identified through deep sequence analysis and more than 800 cases reported in Wuhan and various cases reported in Japan, Thailand, South Korea and the USA. This novel coronavirus was discovered by NHC Key Laboratory of System Biology and Christophe Meraux, China that was confirmed by the WHO[10].WHO declared the public health risks about coronavirus in January 2020 and also gave information that there is no vaccine approved till now[9]. It is confirmed through phylogenetic analysis that the origin of Coronavirus is a bat that transmitted human to human by droplets/air. Infected persons have symptoms such as diarrhea, thrombocytopenia, fever, and respiratory disorders. Further analysis has been made by Real Time-PCR for the gene coding internal RNA-RNA dependent polymerase and Spike’s receptor- binding domain and these analyses after that conformed through Sanger Sequencing and full genome analysis is done by Next Generation Sequencing(NGS)[11-13].The mentioned nonstructural proteins such as PLpro, RNA-RNA dependent polymerase and chymotrypsin-like protease are conserved among all types of coronaviruses, therefore, these proteins could be used as strong antiviral targets to develop a vaccine against SARS and MERS[13].
At present time, no vaccine is availableto cure coronavirus infection in humans. Traditionally two medicines such as ShuFengJieDu and Lianhuaqingwen capsules are usedto treat COVID-19 in China [14]. Many other options could be used for the treatment of SARS-CoV-2 such as vaccines, peptides, monoclonal-oligonucleotides based therapies, and small molecule drugs but these preventive methods need months to years for the development [15].
The purpose of immunization is to design vaccines that have to initiate rapid immune response against pathogen’s antigen. In Vitro vaccine designing could be expensive,time exhausting and there is a need to culture the viruses which is not safe biologically[16]. In immunoinformatic,there is no need to culture viruses and it is biologically safe.B-cells and T- cells are important cells of the immune system that produce immune responses. Immunoinformatics is sub-field of bioinformatics that consist of many bioinformatics tools and databases. Immunological data prediction have been done with the help of these tools. In the advancement of tools, now it has been easy for the scientists to predict B-cell and T-cell epitopes [17] for the development of vaccine sub-unit construct [18, 19]. The basic mechanism to design the peptide vaccine is the prediction of the B-celland T-cell epitopes that are antigenic having particular immune-responses [20]. Therapeutic strategies for the treatment of the E-bola, Zika, and MERS-COV viruses have been designed by the use ofthe in silico peptide prediction approach in various studies [21-23]. The recent study was designed to predict epitopes from SARS-COV-2 PLpro which may be helpful to develop vaccine in future.
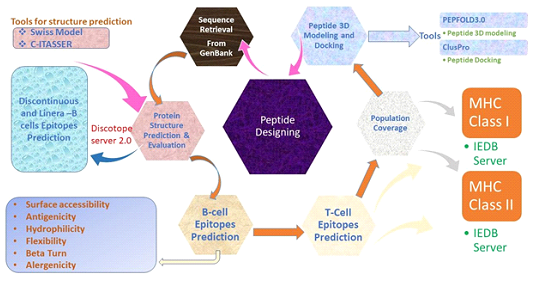
2. Methodology
2.1 Protein structure analysis/Physiochemical Properties prediction of PLpro
The protein sequence of PLpro was taken from Genbank(ID: QHD43415.1)[24]. To evaluate the chemical and physical properties of PLpro the Expasy ProtParam tool was used [25]. GOR4 was used to analyze the protein’s secondary structure. Protein’s transmembrane topology was analyzed usingthe TMHMM tool. Di-sulphide-bonds of the protein were forecasted by DIANNA v1.1[26]. Antigenicity and Allergenicity of protein werepredicted viaVAXIJEN & ALLERTOP servers [27, 28].
2.2 3D Structure Prediction& Validation
The tertiary structure of the PLpro was identified through different online tools like Phyre 2,Swissmodel, C-I tasser and Raptor x[29-31]. Galaxy refine server was used to refine the retrieved model [32]. The refined structure was then checked in PROSA web server that offers a quality factor. The quality-factor suggests a potential protein structure defect, beyond the usual range of native proteins [33]. Ramachandran plot was created with the PROCHECK principle via rampage server to validate the refined structure [34]. To evaluate the details of unbounded interactions, ERRAT server was used [35].
2.3 Prediction of B-cell epitopes
B-cell epitopes help in defending the immune-system. ABC-PRED was applied to identify B- cell epitopes with the threshold value of 0.51[36]. Epitopes visible on the surface were selected for antigenicity testing via VaxiJen server at 0.5 threshold. Kolaskar and Tongaonkar Antigenicity scale, Parkar hydrophilicity prediction algorithm, Karplus and Schulz flexibility prediction tool, Emini surface accessibility prediction method were applied to execute antigenicity, hydrophilicity, flexibility, surface accessibility analysis respectively [37]. As discontinuous epitopes have higher dominant properties than linear epitopes and are more evident.Discontinuous epitopes were identified through DiscoTope 2.0server using three- dimensional structure of PLpro protein [38]. Pymol was applied to visualize the location of epitopes on the 3D protein structure [39].
2.4 T-cell epitopes prediction
T-cell epitopes have avital role in designing of a vaccine. MHC-I &MHCII T cell epitopes were predicted through the IEDB database having a length of peptides 12 and 15 mer respectively[40].HLA alleles were chosen in large numbers so that epitope prediction should be accurate. For prediction, all the alleles were selected and the sequence was given in FASTA format. T-cell epitopes having lower ranking score which is less than 2.0 considered good binders and could be used for further research.
2.5 Conservation Analysis of Epitopes
Conservation analysis was done by the IEDB server database. Epitopes with 100% conservation were selected[41].
2.6 Population Coverage Estimation
The distribution and expression of HLA alleles dependupon the world’s regions and ethnicities, therefore impacting the efficient production of an epitope-based vaccine [42]. In this study selected MHC class I and MHC class II epitopes and related HLA alleles were used for population coverage analisis via IEDB population coverage tool [43]. Population coverage of each epitope was estimated in the regions where SARS-COV-2 cases were reported.
2.7 Properties Evaluation of Peptides
Expassy Protparam tool was applied to check the physiochemical properties of peptides like molecular weight and theoretical pI. VaxiJen v 2.0 and Allergen Fp 1.0 was used to forecast Antigenicity and Allergenicity of the peptide [44]. Toxic and non-toxic peptide prediction was done by ToxinPred but for further analysis, non-toxic peptide was selected [45]. To forecast peptide digester enzymes; the protein-digesting enzyme server was used.
2.8 Peptides Modeling and Docking
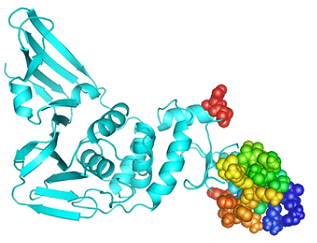
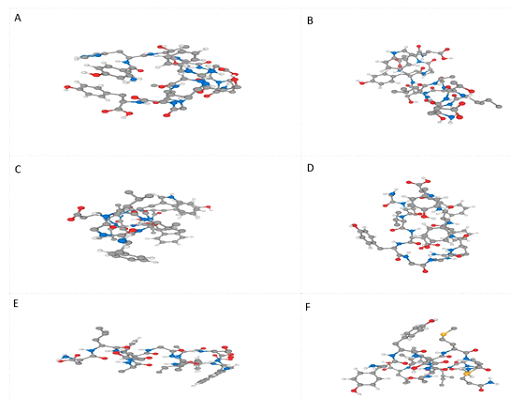
PEPFOLD was applied for the prediction of tertiary structures of six MHC I epitopes. PEPFOLD creates five models for each peptide and the best model was picked for further work [46]. 3D structure of the HLA –B7 allele was downloaded from PDB (ID: 3VCL).ClusPro (v.2) was used for molecular docking [47].
3. Results
3.1 Sequence retrieval & structural analysis
ProtParam was used to evaluate physiochemical properties of SARS COV-2 PLpro. Protein is in stable form, acidic and hydrophilic and has molecular weight 217252.61 kDa. Antigenicity and allergenicity of this sequence weretested by AllerTop and VaxiJen respectively.The results indicate thenon-allergenic and antigenic nature of proteinhaving an antigenicity value of0.5142. DIANNA v1.1 was used to evaluate 51 disulphide bonds of protein (Additional file 1: Table S1). Four transmembrane helices were predicted through TMHMM. Residues of protein that were on transmembrane helix were from 1413-1435, 1500-1522, 1532-1554 and 1561-1583. Residues from 1-1412, 1523-1531 and 1584-1945 were found on the outer surface. Protein residues from 1436-1499 and 1555-1560 were found in the inner surface of the core region. GOR4was used to forecast the details of the secondary structure of the target protein. Among the 1945 amino acids, the formation of α-helix consists of 508 amino acids representing 26.12%, 472 in beta strands representing 24.27% and 965 amino acids forms coils representing 49.61% of the total protein composition.
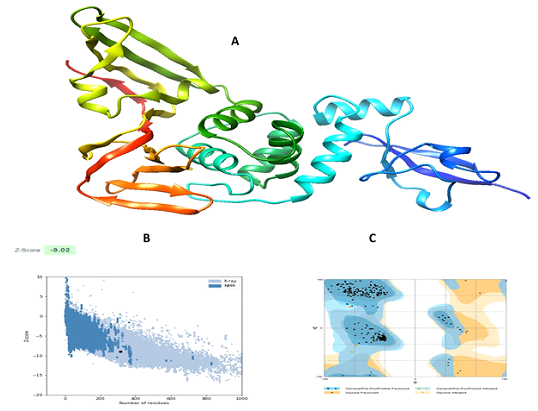
3.2 Tertiary Structure Prediction and Validation
Models retrieved from the Swiss model, C-I Tasser, Raptor x and Phyre 2 were evaluated by Ramachandran plot analysis.The model retrieved from the Swiss Model was best compared to other resources.The protein homology model based on the quality of QMEAN was developed in the Swiss Model. It providesQMean value and GMQE(Global Model Quality Estimation) value.GMQE represents the certainty of a model considering the coverage, arrangement of the target and the template.Higher GMQE value represents a better quality of the model. It is usually measuredin the range of 0 and 1.GMQE of the model was 0.11 and QMean was -0.28 which indicates the high quality of the protein structure. The best template was found to be 3e9s having a sequence identity of 82.86%(Figure 1a).The model was then refined by the Galaxy refine server.The improved model has 98.1% favored region in RAMPAGE and poor rotamers as 0.4%, qrmsd as 0.298, clash score as 10.6, MolProbity as 1.542(Figure 1c). ProSA-web also gives−9.02 Z, which is within an acceptable range (Figure 1b). These results show that the refined model has goodquality.
3.3 B cell epitopes prediction
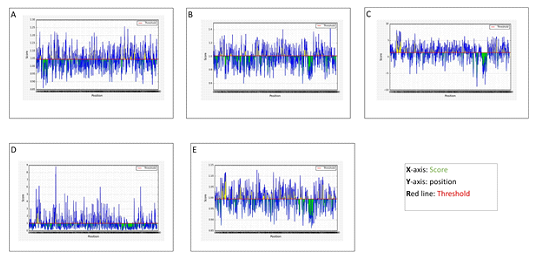
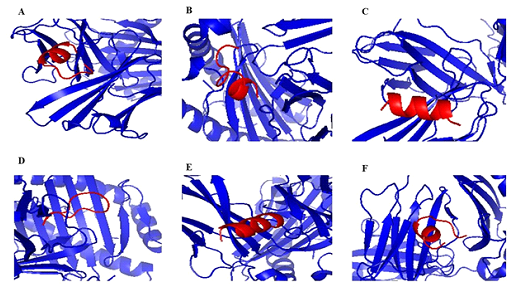
B cell epitope of target protein was checked by ABCpred and TMHMM server were used to check surface availabity. VaxiJen was used to find the antigenicity of the epitopes. Epitopes that were 100% conserved, evident on the surface and antigenic were picked. A Total of 53 epitopes was chosen. ‘KPLEFGATSAALQP’ and ‘EDDYQGKPLEFGAT’ had high Antigenicity (1.7) and score(Table 1). We used Pymol to visualize the position of epitopes on the protein(Figure 2). Besides, surface accessibility of B cell epitopes was assessed. Through observing their concentration in previously identified B cell epitopes and physiochemical properties, the target protein was evaluatedfor prediction of B cell epitopes by Kolaskar and Tongaonkar antigenicity tool.Window size and a threshold werekept at 7 and 1.045 respectively. The antigenic propensity value of PLpro was predicted as 1.041 Average, 0.859 Minimum and 1.258 Maximum(Figure 3A).Experimental research reveals that antibodies or alleles binding section of epitopes is elastic. Hydrophilic protein areas have a role in triggering an immune response and usually have surface exposure.Vaxijen antigenic value and the ABCpred score indicate that all predicted epitopes belong to the extracellular area of transmembrane proteins, which might boost a host defensive reaction during SARS-COV-2 infection. The parker hydrophilicity prediction algorithm with a threshold of 1.279 was used to evaluate hydrophilicity.Values calculated by parker hydrophilicity were 1.409Average, -8.971 Minimum and 8.129 Maximum(Figure 3C). Emini surface accessibility prediction method with a threshold of 1.009 was used to calculate the surface abundance of potential B cell epitopes. Additional file 2 (Table S2)contains the findings of the Emini surface accessibility(Figure 3D). Chou and Fasman beta-turn analyzing algorithm was used for the prediction of beta-turn because beta-turns play a vital role in initiating the defensive response. Beta-turns are hydrophilic and exposed to the surface. It calculated the values of 0.973Average, 0.593Minimum and 1.414 Maximum at the threshold of 1.009. The results show that the area from 1098-1104 amino acids and 1849-1855 amino acids are more likely to persuade beta turns in peptide structure(Figure 3B).Flexibility analysis by Karplus and schulz tool showed that the area from673-679 amino acids is highly versatile (Figure 3E).
DiscoTope 2.0 server was applied to trackthe surface abundance of residual contact numbers and use the novel amino acid score to preview discontinuous epitopes to further improve the specificity and variety of B cell epitopes.At − 3.700 threshold, 22.000 Angstroms propensity score radius and 90% specificity, twenty-nine discontinuous epitopes were identified from the 3D structure of the protein(Table 2). Pymol visualized the location of epitopes on the 3D protein structure(Figure 4).
Table 1: Emini Surface accessibility of PLpro predicted by EMINI surface accessibility tool
Figure 3: (A) Prediction of antigenic determinants of PLpro protein using Kolaskar and Tongaonkar Antigenicity Scale; (B) Beta turns analyses in PLpro protein using Chou and Fasman Beta Turn Prediction; (C) Hydrophilicity Prediction of using PLpro protein Parker Hydrophilicity; (D) Surface Accessibility Analyses of PLpro protein using Emini Surface Accessibility Scale; (E) Flexibility Analyses of PLpro protein using Karplus and Schulz Flexibility Scale.
Table 2: Discontinuous epitopes predicted through DiscoTop 2.0 server
|
Residues Position |
Residues Name |
Number of Contacts |
Propensity Score |
Discotop Score |
|
746 |
GLU |
4 |
2.165 |
1.456 |
|
747 |
VAL |
3 |
1.617 |
1.086 |
|
748 |
ARG |
12 |
1.266 |
-0.26 |
|
749 |
THR |
8 |
0.303 |
-0.652 |
|
750 |
ILE |
19 |
-0.128 |
-2.378 |
|
751 |
LYS |
3 |
-0.5 |
-0.788 |
|
763 |
THR |
3 |
-3.163 |
-3.144 |
|
765 |
VAL |
3 |
-0.715 |
-0.977 |
|
767 |
ASP |
10 |
0.338 |
-0.85 |
|
768 |
MET |
12 |
1.024 |
-0.474 |
|
769 |
SER |
3 |
1.2 |
0.717 |
|
770 |
MET |
9 |
1.122 |
-0.042 |
|
771 |
THR |
13 |
0.48 |
-1.07 |
|
773 |
GLY |
10 |
-0.637 |
-1.714 |
|
774 |
GLN |
11 |
0.154 |
-1.129 |
|
775 |
GLN |
26 |
-0.562 |
-3.487 |
|
787 |
THR |
10 |
-1.994 |
-2.915 |
|
788 |
LYS |
2 |
-0.051 |
-0.275 |
|
789 |
ILE |
11 |
0.306 |
-0.994 |
|
790 |
LYS |
8 |
0.659 |
-0.337 |
|
791 |
PRO |
28 |
0.942 |
-2.386 |
|
792 |
HIS |
10 |
1.274 |
-0.023 |
|
793 |
ASN |
3 |
1.662 |
1.126 |
|
794 |
SER |
5 |
1.496 |
0.749 |
|
795 |
HIS |
29 |
0.839 |
-2.539 |
|
796 |
GLU |
5 |
-0.023 |
-0.059 |
|
797 |
GLY |
1 |
-0.103 |
-1.012 |
|
806 |
ASP |
8 |
-2.763 |
-3.366 |
|
807 |
ASP |
0 |
-2.535 |
-2.244 |
3.4 T cell epitopes prediction and properties evaluation
IEDB consensus method was used to forecast T-cell MHC Class-I as well as MHC class-II epitopes. Peptides due to strong defensive ability can be bind to multiple alleles therefore; this peptide is considered most appropriate. Their Allergenicity and Antigenicity were checked by Allergen FP 1.0 and VaxiJen respectively and IEDB conservancy tool was used for their conservation analysis of protein sequence. Epitopes that are 100% conserved, highly antigenic, non-allergenicand bound to multiple alleles were screened out. 6 MHC-class-I and 6 MHC class- II alleles were shortlisted. Between MHC class-I epitopes ‘YHTTDPSFLGRY’ has the highest antigenicity (1.3) binding with multiple alleles including HLA-A*01:01HLA-A*26:01, HLA- A*29:02, HLA-A*30:02, HLA-A*25:01, HLA-B*38:01, HLA-A*68:01(Table 3). In MHC class-II epitopes ‘PFVMMSAPPAQYELK’ had higher Antigenicity score(0.8) binding with multiple alleles HLA-DRB1*01:02, HLA-DRB1*09:01, HLA-DRB1*01:01, HLA-DRB1*13:07, HLA-DRB1*04:08, HLA-DRB1*11:07, HLA-DRB1*11:14, HLA-DRB1*13:23, HLA-DRB1*03:05 (Table 4). To estimate peptide digesting enzymes, the protein digesting enzyme server was used. The enzymes that do not digest peptide into fragments is considered a non- digestive enzyme. Peptides that are digestible with multiple enzymes are not stable. Peptides that are digested with fewer enzymes are considered most stable and favorable for vaccine (Table 5).
Table 3: MHC class I binding peptides with their antigenicity score
|
Peptide(position) |
Alleles |
Antigenicity |
|
|
|
MHC Class 1 |
||
|
YHTTDPSFLGRY(25-36) |
HLA-A*01:01, HLA-A*26:01,HLA-A*29:02 HLA-A*30:02, HLA-A*25:01, HLA-B*38:01 HLA-A*68:01 |
1.3 |
|
|
ETISLAGSYKDW (33-44) |
HLA-A*25:01, HLA-A*26:01, HLA-B*57:01 HLA-B*58:01, HLA-A*68:01 |
0.8 |
|
|
FGLVAEWFLAYI(11-22) |
HLA-A*26:01, HLA-A*25:01, HLA-A*29:02 HLA-B*35:01, HLA-A*01:01, HLA-C*08:02 HLA-A*02:01, HLA-B*46:01, HLA-B*15:02 HLA-A*68:02 , HLA-C*12:03 |
0.5 |
|
|
GVVDYGARFYFY(14-25) |
HLA-A*29:02, HLA-A*01:01, HLA-C*05:01 HLA-A*30:02, HLA-A*26:01, HLA-A*11:01 HLA-A*23:01 |
0.7 |
|
|
DLTAFGLVAEWF(7-18) |
HLA-B*46:01, HLA-B*57:01 |
0.6 |
|
|
YYSQLMCQPILL(40-51) |
HLA-A*02:01, HLA-A*24:02, HLA-C*14:02 HLA-C*07:02, HLA-B*48:01, HLA-A*23:01 HLA-B*38:01 |
Table 4: MHC class II binding peptides with their antigenicity score
|
Peptide(position) |
Alleles |
Antigenicity |
|
|
MHC Class 2 |
|
|
SPFVMMSAPPAQYEL(52-66) |
HLA-DRB1*01:01,HLA DRB1*01:02,HLA- DRB1*09:01, HLA-DRB1*13:07,HLA DRB1*04:08,HLA-DRB1*11:07,HLA- DRB1*11:14,HLA-DRB1*13:23, HLA-DRB1*03:05HLA-DRB1*04:04,HLA-DRB1*08:02 |
0.5 |
|
KSAFYILPSIISNEK(20-34) |
HLA-DRB1*04:08, HLA-DRB1*04:26, HLA-DRB1*01:01,HLA-DRB1*11:01, HLA-DRB1*04:21, HLA-DRB1*11:28,HLA-DRB1*13:05, HLA-DRB1*13:07,HLA-DRB1*04:01,HLA-DRB1*13:21 |
0.7 |
|
SAFYILPSIISNEKQ(21-35) |
DRB1*13:07,HLA-DRB1*04:01,HLA-DRB1*13:21 HLA-DRB1*11:01,HLA-DRB1*04:21, HLA-DRB1*11:28, HLA-DRB1*13:05,HLA-DRB1*01:01, HLA- DRB1*13:07, HLA-DRB1*04:01,HLA-DRB1*13:21 |
0.6 |
|
EVITFDNLKTLLSLR(9-23) |
HLA-DRB1*04:01, HLA-DRB1*15:06, HLA-DRB1*04:04 |
0.5 |
|
PFVMMSAPPAQYELK(53-67) |
HLA-DRB1*01:02, HLA-DRB1*09:01, HLA-DRB1*01:01,HLA-DRB1*13:07, HLA-DRB1*04:08, HLA-DRB1*11:07,HLA-DRB1*11:14, HLA- DRB1*13:23, HLA-DRB1*03:05,HLA-DRB1*08:02 |
0.8 |
|
QQESPFVMMSAPPAQ(49-63) |
HLA-DRB1*01:02, HLA-DRB1*01:01, HLA-DRB1*09:01,HLA-DRB1*13:07, HLA-DRB1*04:08, HLA-DRB1*11:07,HLA-DRB1*11:14, HLA-DRB1*13:23,HLA-DRB1*03:05,HLA-DRB1*08:02 |
0.6 |
Table 5: Peptides that are digested with fewer enzymes are considered most stable and favorable for vaccine.
3.5 Population coverage of Peptides
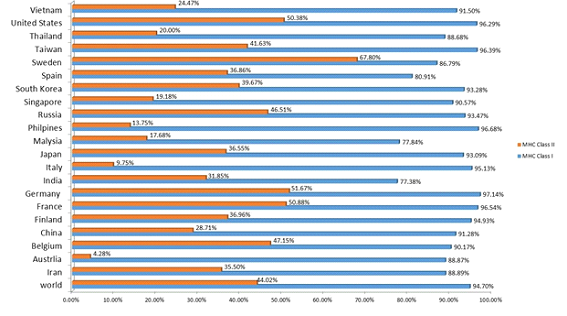
Population coverage was foundforthe selectedsix MHC class-I and six MHC class-II epitopes and related alleles. Epitopes that were selected represent 94.70% and 44.02% of total world population respectively. Highest population coverage of MHC class-I epitopes was found in Germany (97.14 %) and MHC class -II epitopes was found in Sweden (67.80%).SARS-COV-2 MHC class -I epitopes displayed 91.28 % and MHC class -II exhibited 28.71% population coverage in China. Population coverage was higher in those regions where SARS-COV-2 cases were reported (Figure 5).
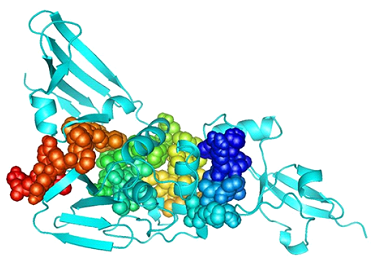
3.6 Molecular Docking
3D structures of selected six MHC class I epitopes were forecasted using PEPFOLD (Figure 6). Molecular docking is necessary to understand the pattern of interaction between protein and peptide. By performing molecular docking the interaction of HLA allele with selected MHC class I epitope was analyzed. To analyze the interaction between allele and peptides, ClusPro (v.2) webserver was used. Rigid docking was performed by the server by sampling billions of conformations, energy minimization and RMSD of the complex [47]. Pymol was used to visualize the docked complexes. Docking results revealed the strong interactions among all selected peptides inside the receptor binding site of HLA allele (Figure 7).
4. Discussion
Several viral infections appeared through many fatal new viruses and these viruses are disastrous for humans. Therefore, vaccine designing to tackle viruses is difficult in a short period [48]. One of the most dangerous infectious viruses such as the 2019 novel coronavirus also called SARS- CoV-2 identified (10 February 2020) in patients of severe pneumonia in Wuhan (China)[15]. Several methods could be used for the treatment of coronavirus such as vaccine, peptides, monoclonal antibodies but we designed peptides for SARS-CoV-2. Genome analysis of 2019- nCoV reveals that four catalytic sites are conserved among all kinds of coronaviruses therefore it is suggested to redesign the already present inhibitors of MERS and SARS[8]. Genomic RNA of SARS-CoV is replicated with the help of replicasepolyproteins that are activated with the help of two enzyme proteases such as 3C-like protease (3CLpro) and Papain like protease (PLpro)[49]. SARS-CoVPLpro replicates the polyprotein by the conserved cleavage sites that are conserved among coronaviruses. Furthermore, it is suggested that aspartic acid (ASP1826) and four cysteine residues are essential to perform the activity of PLpro in SARS-CoV[50]. The vaccine could be beneficial and cost-effective in preventing the viral attack but biologically it takes years to be designed in active form. So at presentimmunoinformatics is used to make peptide-based vaccine which is biologically safe and designed in less time than in-vitro vaccine [51].With the advancement in bioinformatics techniques such as next-generation sequencing(NGS) and proteomics enables us to design effective vaccine bases on peptide through the computational immunology (Immunoinformatics)[52, 53].
The development of the peptide based vaccine was explored in this study targeting the papain- like protease protein of the SARS-COV-2. Primary, secondary, and tertiary structure levels of the protein were analyzed. The primary structure of the protein was analyzed by protparam which indicates that protein was 217252.61 kDa in molecular weight. Protein was acidic according to its theoretical PI value. The calculated aliphatic index and instability index values showed the thermostability and stability of the protein. A negative score of the grand average of hydropathy indicates the hydrophilic nature that renders its interaction with neighboring water molecules. Protein was found to be antigenic, immunogenic and non-allergenic indicating the potential to cause robust immune response without any allergic reactions.
B & T cell epitopes of the target protein were forecasted to assist the immune response of the host.Forecasting T cell &B cell is an important step in vaccine development; therefore both types of epitopes were predicted by the protein sequence and their antigenicity was assessed. ABCpred and Discotope 2.0 servers were used to predict linear and discontinuous B cell epitopes respectively. Different IEDB techniques were used to access solvent accessibility, antigenicity, disulphide bonds, and flexibility. B cell epitopes that were antigenic and present on the outer surface were considered. ‘KPLEFGATSAALQP’ and ‘EDDYQGKPLEFGAT’ had high antigenicity (1.7) and score among B cells. Furthermore, the IEDB consensus approach predicts antigenic T cell determinants with the ability to bind MHC class I and MHC class II. Antigenic, non-allergenic and conserved T cell epitopes that were bound to multiple alleles were considered. YHTTDPSFLGRY’ in MHC class I and ‘PFVMMSAPPAQYELK’ in MHC class II had the highest antigenicity among T cell epitopes. Pymol visualized the location of peptides on the 3D protein structure. Vaccine’s efficacy depends on the population in which the vaccine is being used. MHC class I and MHC class II epitopes that were selected represent 94.70% and 44.02% of the world’s population respectively, indicating vaccine developed would be effective for most of the world population. Allergenicity, toxicity and physiochemical properties of predicted epitopes have also been tested for further improvement in specificity and selection. Digestion analysis tests peptideprotection and stability.Epitopes identified during analysis should be tested for the therapeutic potential for futures studies.
5. Conclusion
A reverse vaccinology approach was implemented in this study to recognize surface-exposed peptides, instead of concentrating on the entire pathogen, an inefficient and effective procedure. B-cells& T-cell epitopes predictedby sequence, conservational, andstructure analysis. B-cell and T- cell epitopes now become vital research due to their predictability, speed, low cost and efficiency. The epitopes with higher antigenicity were selected and these selected peptides would be helpful todesign vaccine against the SARS-COV-2 virus.
References
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395 (2020): 497-506.
- Richman DD, Whitley RJ, Hayden FG. Clinical virology: John Wiley & Sons (2016).
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. New England journal of medicine 348 (2003): 1953-1966.
- Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England journal of medicine 348 (2003): 1967-1976.
- De Groot RJ, Baker SC, Baric RS, et al. Commentary: Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. Journal of virology 87 (2013): 7790-7792.
- Zaki AM, Van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine 367 (2012): 1814-1820.
- xinhuanet.com. [updated 2020-01-23]. Available from: (http://www.xinhuanet.com/english/2020-01/23/c_138728428.htm). .
- De Clercq EJCAAJ. New Nucleoside Analogues for the Treatment of Hemorrhagic Fever Virus Infections. Chem Asian J 14 (2019): 3962-3968.
- Hui DS, I Azhar E, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91 (2020): 264-266.
- Haynes B, Messonnier NE, Cetron MS. First travel-related case of 2019 novel coronavirus detected in United States: press release (2020): 639-3286.
- Chan JF-W, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395 (2020): 514-523.
- Luo G, Gao SJ. Global health concern stirred by emerging viral infections. J Med Virol 92 (2020): 399–400.
- Zumla A, Chan JF, Azhar EI, et al. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov 15 (2016): 327.
- Lu HJBt. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 14 (2020): 69-71.
- Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Publishing Group 19 (2020): 149-150.
- Lo Y-T, Pai T-W, Wu W-K, et al. Prediction of conformational epitopes with the use of a knowledge-based energy function and geometrically related neighboring residue characteristics. BMC Bioinformatics 14 (2013): 3.
- Yang X, Yu XJ. An introduction to epitope prediction methods and software. Rev Med Virol 19 (2009): 77- 96.
- De Gregorio E, Rappuoli R. Vaccines for the future: learning from human immunology. Microb Biotechnol 5 (2012): 149-155.
- Patronov A, Doytchinova IJOb. T-cell epitope vaccine design by immunoinformatics. Open Biol 3 (2013): 120139.
- Abdelmageed MI, Abdelmoneim AH, Mustafa MI, et al. Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV: An immunoinformatics approach. Biomed Res Int 2020 (2020).
- Ahmad B, Ashfaq UA, Rahman M-u, et al. Conserved B and T cell epitopes prediction of ebola virus glycoprotein for vaccine development: an immuno-informatics approach. Microbial pathogenesis 132 (2019): 243-253.
- Ashfaq UA, Ahmed B. De novo structural modeling and conserved epitopes prediction of Zika virus envelop protein for vaccine development. Viral immunology 29 (2016): 436-443.
- Ul Qamar MT, Saleem S, Ashfaq UA, et al. Epitope-based peptide vaccine design and target site depiction against Middle East Respiratory Syndrome Coronavirus: an immune- informatics study. Journal of translational medicine 17 (2019): 362.
- Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic acids research 37 (2008): 26-31.
- Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic acids research 31 (2003): 3784-3788.
- Ferrè F, Clote P. DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic acids research 34 (2006): 182-185.
- Dimitrov I, Flower DR, Doytchinova I, et al. AllerTOP-a server for in silico prediction of allergens. BMC bioinformatics 20 (2013).
- Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC bioinformatics 8 (2007): 4.
- Peng J, Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins: Structure, Function, and Bioinformatics. Proteins 79 (2011): 161-171.
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic acids research 46 (2018): 296- 303.
- Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols 10 (2015): 845.
- Shin W-H, Lee GR, Heo L, et al. Prediction of protein structure and interaction by GALAXY protein modeling programs. Bio Design 2 (2014): 1-11.
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research 35 (2007): 407-410.
- Lovell SC, Davis IW, Arendall III WB, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Bioinformatics 50 (2003): 437-450.
- Gundampati RK, Chikati R, Kumari M, et al. Protein- protein docking on molecular models of Aspergillus niger RNase and human actin: novel target for anticancer therapeutics. J Mol Model 18 (2012): 653-662.
- Saha S, Raghava GPS. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: Structure, Function, and Bioinformatics. J Mol Model 65 (2006): 40-48.
- Jespersen MC, Peters B, Nielsen M, et al. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic acids research 45 (2017): 24-29.
- Sun P, Ju H, Liu Z, et al. Bioinformatics resources and tools for conformational B-cell epitope prediction. Computational and mathematical methods in medicine. Comput Math Methods Med 2013 (2013).
- DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newsletter on protein crystallography. CCP4 Newsletter Pro. Crystallogr 40 (2002): 82-92.
- Wang P, Sidney J, Kim Y, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC bioinformatics 11 (2010): 568.
- Bui H-H, Sidney J, Li W, et al. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC bioinformatics 8 (2007): 361.
- Adhikari UK, Tayebi M, Rahman MM. Immunoinformatics approach for epitope-based peptide vaccine design and active site prediction against polyprotein of emerging oropouche virus. Journal of immunology research 2018 (2018).
- Bui H-H, Sidney J, Dinh K, et al. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC bioinformatics 7 (2006): 153.
- Dimitrov I, Naneva L, Doytchinova I, et al. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30 (2014): 846-851.
- Gupta S, Kapoor P, Chaudhary K, et al. In silico approach for predicting toxicity of peptides and proteins. PloS one 8 (2013).
- Lamiable A, Thévenet P, Rey J, et al. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic acids research 44 (2016): 449-454.
- Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein–protein docking. Nature protocols 12 (2017): 255.
- Adhikari UK, Tayebi M, Rahman MM. Immunoinformatics approach for epitope-based peptide vaccine design and active site prediction against polyprotein of emerging oropouche virus. Journal of Immunology Research 2018 (2018): 1-22
- Baker SC, Shieh C, Soe LH, et al. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol 63 (1989): 3693-3699.
- Goldsmith CS, Tatti KM, Ksiazek TG, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis 10 (2004): 320–326.
- María R, Arturo C, Alicia JA, et al. The impact of bioinformatics on vaccine design and development: InTech, Rijeka, Croatia (2017).
- Groot ASD, Rappuoli RJErov. Genome-derived vaccines. Expert Rev Vaccines 3 (2004): 59-76.
- De Groot AS. Immunome-derived vaccines. Taylor & Francis 4 (2004): 767-772.
Supplementary Materials
Table S1: Disulphide bonds of PLpro predicted by DIANNA v 1.1
|
Cysteine sequence position |
Distance |
Bond |
Score |
|
39 - 55 |
16 |
VLNEKCSAYTV-VNEFACVVADA |
0.01058 |
|
39 - 104 |
65 |
VLNEKCSAYTV-ASHMYCSFYPP |
0.03525 |
|
39 - 118 |
79 |
VLNEKCSAYTV-EEEGDCEEEEF |
0.01323 |
|
39 - 285 |
246 |
VLNEKCSAYTV-KVGGSCVLSGH |
0.0104 |
|
39 - 296 |
257 |
VLNEKCSAYTV-NLAKHCLHVVG |
0.01043 |
|
39 - 347 |
308 |
VLNEKCSAYTV-HSLRVCVDTVR |
0.01101 |
|
39 - 417 |
378 |
VLNEKCSAYTV-KKIKACVEEVT |
0.01055 |
|
39 - 531 |
492 |
VLNEKCSAYTV-TVLKKCKSAFY |
0.01039 |
|
39 - 574 |
535 |
VLNEKCSAYTV-KLMPVCVETKA |
0.01042 |
|
39 - 856 |
817 |
VLNEKCSAYTV-WADNNCYLATA |
0.0104 |
|
39 - 893 |
854 |
VLNEKCSAYTV-EAANFCALILA |
0.01054 |
|
39 - 900 |
861 |
VLNEKCSAYTV-LILAYCNKTVG |
0.0345 |
|
39 - 926 |
887 |
VLNEKCSAYTV-ANLDSCKRVLN |
0.01045 |
|
39 - 934 |
895 |
VLNEKCSAYTV-VLNVVCKTCGQ |
0.0104 |
|
39 - 937 |
898 |
VLNEKCSAYTV-VVCKTCGQQQT |
0.86264 |
|
39 - 969 |
930 |
VLNEKCSAYTV-GVQIPCTCGKQ |
0.0104 |
|
39 - 971 |
932 |
VLNEKCSAYTV-QIPCTCGKQAT |
0.01043 |
|
39 - 1005 |
966 |
VLNEKCSAYTV-HGTFTCASEYT |
0.01662 |
|
39 - 1015 |
976 |
VLNEKCSAYTV-TGNYQCGHYKH |
0.01051 |
|
39 - 1029 |
990 |
VLNEKCSAYTV-KETLYCIDGAL |
0.01066 |
|
39 - 1071 |
1032 |
VLNEKCSAYTV-LDGVVCTEIDP |
0.01513 |
|
39 - 1114 |
1075 |
VLNEKCSAYTV-NFKFVCDNIKF |
0.01067 |
|
39 - 1191 |
1152 |
VLNEKCSAYTV-KPNTWCIRCLW |
0.01048 |
|
39 - 1194 |
1155 |
VLNEKCSAYTV-TWCIRCLWSTK |
0.03167 |
|
39 - 1223 |
1184 |
VLNEKCSAYTV-MDNLACEDLKP |
0.01079 |
|
39 - 1246 |
1207 |
VLNEKCSAYTV-KDVLECNVKTT |
0.01381 |
|
39 - 1342 |
1303 |
VLNEKCSAYTV-NIVTRCLNRVC |
0.01037 |
|
39 - 1347 |
1308 |
VLNEKCSAYTV-CLNRVCTNYMP |
0.01037 |
|
39 - 1362 |
1323 |
VLNEKCSAYTV-LLLQLCTFTRS |
0.01038 |
|
39 - 1392 |
1353 |
VLNEKCSAYTV-SVGKFCLEASF |
0.01037 |
|
39 - 1421 |
1382 |
VLNEKCSAYTV-LLLSVCLGSLI |
0.01037 |
|
39 - 1445 |
1406 |
VLNEKCSAYTV-GMPSYCTGYRE |
0.01038 |
|
39 - 1464 |
1425 |
VLNEKCSAYTV-TIATYCTGSIP |
0.9982 |
|
39 - 1470 |
1431 |
VLNEKCSAYTV-TGSIPCSVCLS |
0.02564 |
|
39 - 1473 |
1434 |
VLNEKCSAYTV-IPCSVCLSGLD |
0.02302 |
|
39 - 1586 |
1547 |
VLNEKCSAYTV-HVVDGCNSSTC |
0.01615 |
|
39 - 1591 |
1552 |
VLNEKCSAYTV-CNSSTCMMCYK |
0.01084 |
|
39 - 1594 |
1555 |
VLNEKCSAYTV-STCMMCYKRNR |
0.03241 |
|
39 - 1605 |
1566 |
VLNEKCSAYTV-ATRVECTTIVN |
0.01042 |
|
39 - 1627 |
1588 |
VLNEKCSAYTV-GGKGFCKLHNW |
0.01072 |
|
39 - 1634 |
1595 |
VLNEKCSAYTV-LHNWNCVNCDT |
0.06308 |
|
39 - 1637 |
1598 |
VLNEKCSAYTV-WNCVNCDTFCA |
0.0146 |
|
39 - 1641 |
1602 |
VLNEKCSAYTV-NCDTFCAGSTF |
0.01845 |
|
39 - 1731 |
1692 |
VLNEKCSAYTV-DGKSKCEESSA |
0.0104 |
|
39 - 1748 |
1709 |
VLNEKCSAYTV-YSQLMCQPILL |
0.01074 |
|
39 - 1836 |
1797 |
VLNEKCSAYTV-KDVVECLKLSH |
0.01049 |
|
39 - 1852 |
1813 |
VLNEKCSAYTV-VTGDSCNNYML |
0.01474 |
|
39 - 1873 |
1834 |
VLNEKCSAYTV-RDLGACIDCSA |
0.01037 |
|
39 - 1876 |
1837 |
VLNEKCSAYTV-GACIDCSARHI |
0.0229 |
|
39 - 1926 |
1887 |
VLNEKCSAYTV-PFKLTCATTRQ |
0.01038 |
|
55 - 104 |
49 |
VNEFACVVADA-ASHMYCSFYPP |
0.01125 |
|
55 - 118 |
63 |
VNEFACVVADA-EEEGDCEEEEF |
0.03137 |
|
55 - 285 |
230 |
VNEFACVVADA-KVGGSCVLSGH |
0.01051 |
|
55 - 296 |
241 |
VNEFACVVADA-NLAKHCLHVVG |
0.01039 |
|
55 - 347 |
292 |
VNEFACVVADA-HSLRVCVDTVR |
0.01046 |
|
55 - 417 |
362 |
VNEFACVVADA-KKIKACVEEVT |
0.01073 |
|
55 - 531 |
476 |
VNEFACVVADA-TVLKKCKSAFY |
0.01069 |
|
55 - 574 |
519 |
VNEFACVVADA-KLMPVCVETKA |
0.01113 |
|
55 - 856 |
801 |
VNEFACVVADA-WADNNCYLATA |
0.01164 |
|
55 - 893 |
838 |
VNEFACVVADA-EAANFCALILA |
0.0104 |
|
55 - 900 |
845 |
VNEFACVVADA-LILAYCNKTVG |
0.01049 |
|
55 - 926 |
871 |
VNEFACVVADA-ANLDSCKRVLN |
0.01047 |
|
55 - 934 |
879 |
VNEFACVVADA-VLNVVCKTCGQ |
0.01059 |
|
55 - 937 |
882 |
VNEFACVVADA-VVCKTCGQQQT |
0.01414 |
|
55 - 969 |
914 |
VNEFACVVADA-GVQIPCTCGKQ |
0.01045 |
|
55 - 971 |
916 |
VNEFACVVADA-QIPCTCGKQAT |
0.0109 |
|
55 - 1005 |
950 |
VNEFACVVADA-HGTFTCASEYT |
0.0132 |
|
55 - 1015 |
960 |
VNEFACVVADA-TGNYQCGHYKH |
0.02341 |
|
55 - 1029 |
974 |
VNEFACVVADA-KETLYCIDGAL |
0.01089 |
|
55 - 1071 |
1016 |
VNEFACVVADA-LDGVVCTEIDP |
0.01213 |
|
55 - 1114 |
1059 |
VNEFACVVADA-NFKFVCDNIKF |
0.01347 |
|
55 - 1191 |
1136 |
VNEFACVVADA-KPNTWCIRCLW |
0.01041 |
|
55 - 1194 |
1139 |
VNEFACVVADA-TWCIRCLWSTK |
0.01044 |
|
55 - 1223 |
1168 |
VNEFACVVADA-MDNLACEDLKP |
0.01052 |
|
55 - 1246 |
1191 |
VNEFACVVADA-KDVLECNVKTT |
0.01056 |
|
55 - 1342 |
1287 |
VNEFACVVADA-NIVTRCLNRVC |
0.01038 |
|
55 - 1347 |
1292 |
VNEFACVVADA-CLNRVCTNYMP |
0.01037 |
|
55 - 1362 |
1307 |
VNEFACVVADA-LLLQLCTFTRS |
0.24573 |
|
55 - 1392 |
1337 |
VNEFACVVADA-SVGKFCLEASF |
0.01041 |
|
55 - 1421 |
1366 |
VNEFACVVADA-LLLSVCLGSLI |
0.01384 |
|
55 - 1445 |
1390 |
VNEFACVVADA-GMPSYCTGYRE |
0.01047 |
|
55 - 1464 |
1409 |
VNEFACVVADA-TIATYCTGSIP |
0.01861 |
|
55 - 1470 |
1415 |
VNEFACVVADA-TGSIPCSVCLS |
0.03859 |
|
55 - 1473 |
1418 |
VNEFACVVADA-IPCSVCLSGLD |
0.01047 |
|
55 - 1586 |
1531 |
VNEFACVVADA-HVVDGCNSSTC |
0.01054 |
|
55 - 1591 |
1536 |
VNEFACVVADA-CNSSTCMMCYK |
0.01043 |
|
55 - 1594 |
1539 |
VNEFACVVADA-STCMMCYKRNR |
0.01251 |
|
55 - 1605 |
1550 |
VNEFACVVADA-ATRVECTTIVN |
0.01037 |
|
55 - 1627 |
1572 |
VNEFACVVADA-GGKGFCKLHNW |
0.01146 |
|
55 - 1634 |
1579 |
VNEFACVVADA-LHNWNCVNCDT |
0.01066 |
|
55 - 1637 |
1582 |
VNEFACVVADA-WNCVNCDTFCA |
0.01171 |
|
55 - 1641 |
1586 |
VNEFACVVADA-NCDTFCAGSTF |
0.99999 |
|
55 - 1731 |
1676 |
VNEFACVVADA-DGKSKCEESSA |
0.01042 |
|
55 - 1748 |
1693 |
VNEFACVVADA-YSQLMCQPILL |
0.02261 |
|
55 - 1836 |
1781 |
VNEFACVVADA-KDVVECLKLSH |
0.01049 |
|
55 - 1852 |
1797 |
VNEFACVVADA-VTGDSCNNYML |
0.82462 |
|
55 - 1873 |
1818 |
VNEFACVVADA-RDLGACIDCSA |
0.01037 |
|
55 - 1876 |
1821 |
VNEFACVVADA-GACIDCSARHI |
0.01106 |
|
55 - 1926 |
1871 |
VNEFACVVADA-PFKLTCATTRQ |
0.01038 |
|
104 - 118 |
14 |
ASHMYCSFYPP-EEEGDCEEEEF |
0.01038 |
|
104 - 285 |
181 |
ASHMYCSFYPP-KVGGSCVLSGH |
0.01148 |
|
104 - 296 |
192 |
ASHMYCSFYPP-NLAKHCLHVVG |
0.01038 |
|
104 - 347 |
243 |
ASHMYCSFYPP-HSLRVCVDTVR |
0.01189 |
|
104 - 417 |
313 |
ASHMYCSFYPP-KKIKACVEEVT |
0.01037 |
|
104 - 531 |
427 |
ASHMYCSFYPP-TVLKKCKSAFY |
0.01037 |
|
104 - 574 |
470 |
ASHMYCSFYPP-KLMPVCVETKA |
0.01037 |
|
104 - 856 |
752 |
ASHMYCSFYPP-WADNNCYLATA |
0.01079 |
|
104 - 893 |
789 |
ASHMYCSFYPP-EAANFCALILA |
0.01038 |
|
104 - 900 |
796 |
ASHMYCSFYPP-LILAYCNKTVG |
0.01236 |
|
104 - 926 |
822 |
ASHMYCSFYPP-ANLDSCKRVLN |
0.01037 |
|
104 - 934 |
830 |
ASHMYCSFYPP-VLNVVCKTCGQ |
0.01037 |
|
104 - 937 |
833 |
ASHMYCSFYPP-VVCKTCGQQQT |
0.01122 |
|
104 - 969 |
865 |
ASHMYCSFYPP-GVQIPCTCGKQ |
0.01038 |
|
104 - 971 |
867 |
ASHMYCSFYPP-QIPCTCGKQAT |
0.01037 |
|
104 - 1005 |
901 |
ASHMYCSFYPP-HGTFTCASEYT |
0.0106 |
|
104 - 1015 |
911 |
ASHMYCSFYPP-TGNYQCGHYKH |
0.01172 |
|
104 - 1029 |
925 |
ASHMYCSFYPP-KETLYCIDGAL |
0.01037 |
|
104 - 1071 |
967 |
ASHMYCSFYPP-LDGVVCTEIDP |
0.01429 |
|
104 - 1114 |
1010 |
ASHMYCSFYPP-NFKFVCDNIKF |
0.01049 |
|
104 - 1191 |
1087 |
ASHMYCSFYPP-KPNTWCIRCLW |
0.01052 |
|
104 - 1194 |
1090 |
ASHMYCSFYPP-TWCIRCLWSTK |
0.02164 |
|
104 - 1223 |
1119 |
ASHMYCSFYPP-MDNLACEDLKP |
0.01041 |
|
104 - 1246 |
1142 |
ASHMYCSFYPP-KDVLECNVKTT |
0.01208 |
|
104 - 1342 |
1238 |
ASHMYCSFYPP-NIVTRCLNRVC |
0.01038 |
|
104 - 1347 |
1243 |
ASHMYCSFYPP-CLNRVCTNYMP |
0.01037 |
|
104 - 1362 |
1258 |
ASHMYCSFYPP-LLLQLCTFTRS |
0.01039 |
|
104 - 1392 |
1288 |
ASHMYCSFYPP-SVGKFCLEASF |
0.01037 |
|
104 - 1421 |
1317 |
ASHMYCSFYPP-LLLSVCLGSLI |
0.01037 |
|
104 - 1445 |
1341 |
ASHMYCSFYPP-GMPSYCTGYRE |
0.01038 |
|
104 - 1464 |
1360 |
ASHMYCSFYPP-TIATYCTGSIP |
0.84832 |
|
104 - 1470 |
1366 |
ASHMYCSFYPP-TGSIPCSVCLS |
0.01055 |
|
104 - 1473 |
1369 |
ASHMYCSFYPP-IPCSVCLSGLD |
0.01088 |
|
104 - 1586 |
1482 |
ASHMYCSFYPP-HVVDGCNSSTC |
0.01065 |
|
104 - 1591 |
1487 |
ASHMYCSFYPP-CNSSTCMMCYK |
0.01037 |
|
104 - 1594 |
1490 |
ASHMYCSFYPP-STCMMCYKRNR |
0.02433 |
|
104 - 1605 |
1501 |
ASHMYCSFYPP-ATRVECTTIVN |
0.01129 |
|
104 - 1627 |
1523 |
ASHMYCSFYPP-GGKGFCKLHNW |
0.63807 |
|
104 - 1634 |
1530 |
ASHMYCSFYPP-LHNWNCVNCDT |
0.01115 |
|
104 - 1637 |
1533 |
ASHMYCSFYPP-WNCVNCDTFCA |
0.01117 |
|
104 - 1641 |
1537 |
ASHMYCSFYPP-NCDTFCAGSTF |
0.01041 |
|
104 - 1731 |
1627 |
ASHMYCSFYPP-DGKSKCEESSA |
0.01038 |
|
104 - 1748 |
1644 |
ASHMYCSFYPP-YSQLMCQPILL |
0.01051 |
|
104 - 1836 |
1732 |
ASHMYCSFYPP-KDVVECLKLSH |
0.01037 |
|
104 - 1852 |
1748 |
ASHMYCSFYPP-VTGDSCNNYML |
0.01053 |
|
104 - 1873 |
1769 |
ASHMYCSFYPP-RDLGACIDCSA |
0.01037 |
|
104 - 1876 |
1772 |
ASHMYCSFYPP-GACIDCSARHI |
0.0113 |
|
104 - 1926 |
1822 |
ASHMYCSFYPP-PFKLTCATTRQ |
0.01037 |
|
118 - 285 |
167 |
EEEGDCEEEEF-KVGGSCVLSGH |
0.0104 |
|
118 - 296 |
178 |
EEEGDCEEEEF-NLAKHCLHVVG |
0.01038 |
|
118 - 347 |
229 |
EEEGDCEEEEF-HSLRVCVDTVR |
0.01037 |
|
118 - 417 |
299 |
EEEGDCEEEEF-KKIKACVEEVT |
0.01044 |
|
118 - 531 |
413 |
EEEGDCEEEEF-TVLKKCKSAFY |
0.0104 |
|
118 - 574 |
456 |
EEEGDCEEEEF-KLMPVCVETKA |
0.01038 |
|
118 - 856 |
738 |
EEEGDCEEEEF-WADNNCYLATA |
0.01063 |
|
118 - 893 |
775 |
EEEGDCEEEEF-EAANFCALILA |
0.01037 |
|
118 - 900 |
782 |
EEEGDCEEEEF-LILAYCNKTVG |
0.01037 |
|
118 - 926 |
808 |
EEEGDCEEEEF-ANLDSCKRVLN |
0.01037 |
|
118 - 934 |
816 |
EEEGDCEEEEF-VLNVVCKTCGQ |
0.01037 |
|
118 - 937 |
819 |
EEEGDCEEEEF-VVCKTCGQQQT |
0.01038 |
|
118 - 969 |
851 |
EEEGDCEEEEF-GVQIPCTCGKQ |
0.0104 |
|
118 - 971 |
853 |
EEEGDCEEEEF-QIPCTCGKQAT |
0.01039 |
|
118 - 1005 |
887 |
EEEGDCEEEEF-HGTFTCASEYT |
0.01038 |
|
118 - 1015 |
897 |
EEEGDCEEEEF-TGNYQCGHYKH |
0.0121 |
|
118 - 1029 |
911 |
EEEGDCEEEEF-KETLYCIDGAL |
0.01047 |
|
118 - 1071 |
953 |
EEEGDCEEEEF-LDGVVCTEIDP |
0.01038 |
|
118 - 1114 |
996 |
EEEGDCEEEEF-NFKFVCDNIKF |
0.01046 |
|
118 - 1191 |
1073 |
EEEGDCEEEEF-KPNTWCIRCLW |
0.01038 |
|
118 - 1194 |
1076 |
EEEGDCEEEEF-TWCIRCLWSTK |
0.01037 |
|
118 - 1223 |
1105 |
EEEGDCEEEEF-MDNLACEDLKP |
0.01082 |
|
118 - 1246 |
1128 |
EEEGDCEEEEF-KDVLECNVKTT |
0.01037 |
|
118 - 1342 |
1224 |
EEEGDCEEEEF-NIVTRCLNRVC |
0.01037 |
|
118 - 1347 |
1229 |
EEEGDCEEEEF-CLNRVCTNYMP |
0.01037 |
|
118 - 1362 |
1244 |
EEEGDCEEEEF-LLLQLCTFTRS |
0.01037 |
|
118 - 1392 |
1274 |
EEEGDCEEEEF-SVGKFCLEASF |
0.01037 |
|
118 - 1421 |
1303 |
EEEGDCEEEEF-LLLSVCLGSLI |
0.01037 |
|
118 - 1445 |
1327 |
EEEGDCEEEEF-GMPSYCTGYRE |
0.01038 |
|
118 - 1464 |
1346 |
EEEGDCEEEEF-TIATYCTGSIP |
0.01037 |
|
118 - 1470 |
1352 |
EEEGDCEEEEF-TGSIPCSVCLS |
0.01068 |
|
118 - 1473 |
1355 |
EEEGDCEEEEF-IPCSVCLSGLD |
0.01038 |
|
118 - 1586 |
1468 |
EEEGDCEEEEF-HVVDGCNSSTC |
0.01037 |
|
118 - 1591 |
1473 |
EEEGDCEEEEF-CNSSTCMMCYK |
0.01037 |
|
118 - 1594 |
1476 |
EEEGDCEEEEF-STCMMCYKRNR |
0.01439 |
|
118 - 1605 |
1487 |
EEEGDCEEEEF-ATRVECTTIVN |
0.01038 |
|
118 - 1627 |
1509 |
EEEGDCEEEEF-GGKGFCKLHNW |
0.01145 |
|
118 - 1634 |
1516 |
EEEGDCEEEEF-LHNWNCVNCDT |
0.01037 |
|
118 - 1637 |
1519 |
EEEGDCEEEEF-WNCVNCDTFCA |
0.01059 |
|
118 - 1641 |
1523 |
EEEGDCEEEEF-NCDTFCAGSTF |
0.01456 |
|
118 - 1731 |
1613 |
EEEGDCEEEEF-DGKSKCEESSA |
0.01041 |
|
118 - 1748 |
1630 |
EEEGDCEEEEF-YSQLMCQPILL |
0.0112 |
|
118 - 1836 |
1718 |
EEEGDCEEEEF-KDVVECLKLSH |
0.01038 |
|
118 - 1852 |
1734 |
EEEGDCEEEEF-VTGDSCNNYML |
0.01049 |
|
118 - 1873 |
1755 |
EEEGDCEEEEF-RDLGACIDCSA |
0.01038 |
|
118 - 1876 |
1758 |
EEEGDCEEEEF-GACIDCSARHI |
0.01037 |
|
118 - 1926 |
1808 |
EEEGDCEEEEF-PFKLTCATTRQ |
0.01037 |
|
285 - 296 |
11 |
KVGGSCVLSGH-NLAKHCLHVVG |
0.01078 |
|
285 - 347 |
62 |
KVGGSCVLSGH-HSLRVCVDTVR |
0.01038 |
|
285 - 417 |
132 |
KVGGSCVLSGH-KKIKACVEEVT |
0.01595 |
|
285 - 531 |
246 |
KVGGSCVLSGH-TVLKKCKSAFY |
0.01097 |
|
285 - 574 |
289 |
KVGGSCVLSGH-KLMPVCVETKA |
0.01062 |
|
285 - 856 |
571 |
KVGGSCVLSGH-WADNNCYLATA |
0.01242 |
|
285 - 893 |
608 |
KVGGSCVLSGH-EAANFCALILA |
0.01037 |
|
285 - 900 |
615 |
KVGGSCVLSGH-LILAYCNKTVG |
0.01038 |
|
285 - 926 |
641 |
KVGGSCVLSGH-ANLDSCKRVLN |
0.01066 |
|
285 - 934 |
649 |
KVGGSCVLSGH-VLNVVCKTCGQ |
0.01068 |
|
285 - 937 |
652 |
KVGGSCVLSGH-VVCKTCGQQQT |
0.01038 |
|
285 - 969 |
684 |
KVGGSCVLSGH-GVQIPCTCGKQ |
0.01044 |
|
285 - 971 |
686 |
KVGGSCVLSGH-QIPCTCGKQAT |
0.01098 |
|
285 - 1005 |
720 |
KVGGSCVLSGH-HGTFTCASEYT |
0.01169 |
|
285 - 1015 |
730 |
KVGGSCVLSGH-TGNYQCGHYKH |
0.01446 |
|
285 - 1029 |
744 |
KVGGSCVLSGH-KETLYCIDGAL |
0.01069 |
|
285 - 1071 |
786 |
KVGGSCVLSGH-LDGVVCTEIDP |
0.02752 |
|
285 - 1114 |
829 |
KVGGSCVLSGH-NFKFVCDNIKF |
0.11443 |
|
285 - 1191 |
906 |
KVGGSCVLSGH-KPNTWCIRCLW |
0.01104 |
|
285 - 1194 |
909 |
KVGGSCVLSGH-TWCIRCLWSTK |
0.01037 |
|
285 - 1223 |
938 |
KVGGSCVLSGH-MDNLACEDLKP |
0.01119 |
|
285 - 1246 |
961 |
KVGGSCVLSGH-KDVLECNVKTT |
0.01057 |
|
285 - 1342 |
1057 |
KVGGSCVLSGH-NIVTRCLNRVC |
0.01037 |
|
285 - 1347 |
1062 |
KVGGSCVLSGH-CLNRVCTNYMP |
0.01037 |
|
285 - 1362 |
1077 |
KVGGSCVLSGH-LLLQLCTFTRS |
0.01219 |
|
285 - 1392 |
1107 |
KVGGSCVLSGH-SVGKFCLEASF |
0.01037 |
|
285 - 1421 |
1136 |
KVGGSCVLSGH-LLLSVCLGSLI |
0.01041 |
|
285 - 1445 |
1160 |
KVGGSCVLSGH-GMPSYCTGYRE |
0.01056 |
|
285 - 1464 |
1179 |
KVGGSCVLSGH-TIATYCTGSIP |
0.01038 |
|
285 - 1470 |
1185 |
KVGGSCVLSGH-TGSIPCSVCLS |
0.10547 |
|
285 - 1473 |
1188 |
KVGGSCVLSGH-IPCSVCLSGLD |
0.01055 |
|
285 - 1586 |
1301 |
KVGGSCVLSGH-HVVDGCNSSTC |
0.01044 |
|
285 - 1591 |
1306 |
KVGGSCVLSGH-CNSSTCMMCYK |
0.01043 |
|
285 - 1594 |
1309 |
KVGGSCVLSGH-STCMMCYKRNR |
0.01175 |
|
285 - 1605 |
1320 |
KVGGSCVLSGH-ATRVECTTIVN |
0.0104 |
|
285 - 1627 |
1342 |
KVGGSCVLSGH-GGKGFCKLHNW |
0.5664 |
|
285 - 1634 |
1349 |
KVGGSCVLSGH-LHNWNCVNCDT |
0.0105 |
|
285 - 1637 |
1352 |
KVGGSCVLSGH-WNCVNCDTFCA |
0.01066 |
|
285 - 1641 |
1356 |
KVGGSCVLSGH-NCDTFCAGSTF |
0.13312 |
|
285 - 1731 |
1446 |
KVGGSCVLSGH-DGKSKCEESSA |
0.01164 |
|
285 - 1748 |
1463 |
KVGGSCVLSGH-YSQLMCQPILL |
0.26069 |
|
285 - 1836 |
1551 |
KVGGSCVLSGH-KDVVECLKLSH |
0.01069 |
|
285 - 1852 |
1567 |
KVGGSCVLSGH-VTGDSCNNYML |
0.01603 |
|
285 - 1873 |
1588 |
KVGGSCVLSGH-RDLGACIDCSA |
0.01042 |
|
285 - 1876 |
1591 |
KVGGSCVLSGH-GACIDCSARHI |
0.01106 |
|
285 - 1926 |
1641 |
KVGGSCVLSGH-PFKLTCATTRQ |
0.01044 |
|
296 - 347 |
51 |
NLAKHCLHVVG-HSLRVCVDTVR |
0.01455 |
|
296 - 417 |
121 |
NLAKHCLHVVG-KKIKACVEEVT |
0.01057 |
|
296 - 531 |
235 |
NLAKHCLHVVG-TVLKKCKSAFY |
0.01052 |
|
296 - 574 |
278 |
NLAKHCLHVVG-KLMPVCVETKA |
0.0104 |
|
296 - 856 |
560 |
NLAKHCLHVVG-WADNNCYLATA |
0.01039 |
|
296 - 893 |
597 |
NLAKHCLHVVG-EAANFCALILA |
0.01049 |
|
296 - 900 |
604 |
NLAKHCLHVVG-LILAYCNKTVG |
0.05653 |
|
296 - 926 |
630 |
NLAKHCLHVVG-ANLDSCKRVLN |
0.01062 |
|
296 - 934 |
638 |
NLAKHCLHVVG-VLNVVCKTCGQ |
0.01043 |
|
296 - 937 |
641 |
NLAKHCLHVVG-VVCKTCGQQQT |
0.0829 |
|
296 - 969 |
673 |
NLAKHCLHVVG-GVQIPCTCGKQ |
0.01039 |
|
296 - 971 |
675 |
NLAKHCLHVVG-QIPCTCGKQAT |
0.0105 |
|
296 - 1005 |
709 |
NLAKHCLHVVG-HGTFTCASEYT |
0.0106 |
|
296 - 1015 |
719 |
NLAKHCLHVVG-TGNYQCGHYKH |
0.01059 |
|
296 - 1029 |
733 |
NLAKHCLHVVG-KETLYCIDGAL |
0.01045 |
|
296 - 1071 |
775 |
NLAKHCLHVVG-LDGVVCTEIDP |
0.01467 |
|
296 - 1114 |
818 |
NLAKHCLHVVG-NFKFVCDNIKF |
0.01207 |
|
296 - 1191 |
895 |
NLAKHCLHVVG-KPNTWCIRCLW |
0.01038 |
|
296 - 1194 |
898 |
NLAKHCLHVVG-TWCIRCLWSTK |
0.01335 |
|
296 - 1223 |
927 |
NLAKHCLHVVG-MDNLACEDLKP |
0.0105 |
|
296 - 1246 |
950 |
NLAKHCLHVVG-KDVLECNVKTT |
0.02231 |
|
296 - 1342 |
1046 |
NLAKHCLHVVG-NIVTRCLNRVC |
0.01042 |
|
296 - 1347 |
1051 |
NLAKHCLHVVG-CLNRVCTNYMP |
0.01037 |
|
296 - 1362 |
1066 |
NLAKHCLHVVG-LLLQLCTFTRS |
0.01039 |
|
296 - 1392 |
1096 |
NLAKHCLHVVG-SVGKFCLEASF |
0.01037 |
|
296 - 1421 |
1125 |
NLAKHCLHVVG-LLLSVCLGSLI |
0.01037 |
|
296 - 1445 |
1149 |
NLAKHCLHVVG-GMPSYCTGYRE |
0.01039 |
|
296 - 1464 |
1168 |
NLAKHCLHVVG-TIATYCTGSIP |
0.2882 |
|
296 - 1470 |
1174 |
NLAKHCLHVVG-TGSIPCSVCLS |
0.01239 |
|
296 - 1473 |
1177 |
NLAKHCLHVVG-IPCSVCLSGLD |
0.01309 |
|
296 - 1586 |
1290 |
NLAKHCLHVVG-HVVDGCNSSTC |
0.01114 |
|
296 - 1591 |
1295 |
NLAKHCLHVVG-CNSSTCMMCYK |
0.01038 |
|
296 - 1594 |
1298 |
NLAKHCLHVVG-STCMMCYKRNR |
0.01426 |
|
296 - 1605 |
1309 |
NLAKHCLHVVG-ATRVECTTIVN |
0.01037 |
|
296 - 1627 |
1331 |
NLAKHCLHVVG-GGKGFCKLHNW |
0.01043 |
|
296 - 1634 |
1338 |
NLAKHCLHVVG-LHNWNCVNCDT |
0.01084 |
|
296 - 1637 |
1341 |
NLAKHCLHVVG-WNCVNCDTFCA |
0.01226 |
|
296 - 1641 |
1345 |
NLAKHCLHVVG-NCDTFCAGSTF |
0.56374 |
|
296 - 1731 |
1435 |
NLAKHCLHVVG-DGKSKCEESSA |
0.01075 |
|
296 - 1748 |
1452 |
NLAKHCLHVVG-YSQLMCQPILL |
0.07916 |
|
296 - 1836 |
1540 |
NLAKHCLHVVG-KDVVECLKLSH |
0.0104 |
|
296 - 1852 |
1556 |
NLAKHCLHVVG-VTGDSCNNYML |
0.01081 |
|
296 - 1873 |
1577 |
NLAKHCLHVVG-RDLGACIDCSA |
0.01037 |
|
296 - 1876 |
1580 |
NLAKHCLHVVG-GACIDCSARHI |
0.03981 |
|
296 - 1926 |
1630 |
NLAKHCLHVVG-PFKLTCATTRQ |
0.01038 |
|
347 - 417 |
70 |
HSLRVCVDTVR-KKIKACVEEVT |
0.15833 |
|
347 - 531 |
184 |
HSLRVCVDTVR-TVLKKCKSAFY |
0.07784 |
|
347 - 574 |
227 |
HSLRVCVDTVR-KLMPVCVETKA |
0.012 |
|
347 - 856 |
509 |
HSLRVCVDTVR-WADNNCYLATA |
0.0107 |
|
347 - 893 |
546 |
HSLRVCVDTVR-EAANFCALILA |
0.0105 |
|
347 - 900 |
553 |
HSLRVCVDTVR-LILAYCNKTVG |
0.01075 |
|
347 - 926 |
579 |
HSLRVCVDTVR-ANLDSCKRVLN |
0.0107 |
|
347 - 934 |
587 |
HSLRVCVDTVR-VLNVVCKTCGQ |
0.01082 |
|
347 - 937 |
590 |
HSLRVCVDTVR-VVCKTCGQQQT |
0.01129 |
|
347 - 969 |
622 |
HSLRVCVDTVR-GVQIPCTCGKQ |
0.01052 |
|
347 - 971 |
624 |
HSLRVCVDTVR-QIPCTCGKQAT |
0.01442 |
|
347 - 1005 |
658 |
HSLRVCVDTVR-HGTFTCASEYT |
0.015 |
|
347 - 1015 |
668 |
HSLRVCVDTVR-TGNYQCGHYKH |
0.61951 |
|
347 - 1029 |
682 |
HSLRVCVDTVR-KETLYCIDGAL |
0.01121 |
|
347 - 1071 |
724 |
HSLRVCVDTVR-LDGVVCTEIDP |
0.01203 |
|
347 - 1114 |
767 |
HSLRVCVDTVR-NFKFVCDNIKF |
0.03105 |
|
347 - 1191 |
844 |
HSLRVCVDTVR-KPNTWCIRCLW |
0.01043 |
|
347 - 1194 |
847 |
HSLRVCVDTVR-TWCIRCLWSTK |
0.01107 |
|
347 - 1223 |
876 |
HSLRVCVDTVR-MDNLACEDLKP |
0.48796 |
|
347 - 1246 |
899 |
HSLRVCVDTVR-KDVLECNVKTT |
0.01074 |
|
347 - 1342 |
995 |
HSLRVCVDTVR-NIVTRCLNRVC |
0.0105 |
|
347 - 1347 |
1000 |
HSLRVCVDTVR-CLNRVCTNYMP |
0.01933 |
|
347 - 1362 |
1015 |
HSLRVCVDTVR-LLLQLCTFTRS |
0.01267 |
|
347 - 1392 |
1045 |
HSLRVCVDTVR-SVGKFCLEASF |
0.01096 |
|
347 - 1421 |
1074 |
HSLRVCVDTVR-LLLSVCLGSLI |
0.01045 |
|
347 - 1445 |
1098 |
HSLRVCVDTVR-GMPSYCTGYRE |
0.01058 |
|
347 - 1464 |
1117 |
HSLRVCVDTVR-TIATYCTGSIP |
0.01802 |
|
347 - 1470 |
1123 |
HSLRVCVDTVR-TGSIPCSVCLS |
0.4005 |
|
347 - 1473 |
1126 |
HSLRVCVDTVR-IPCSVCLSGLD |
0.02946 |
|
347 - 1586 |
1239 |
HSLRVCVDTVR-HVVDGCNSSTC |
0.0105 |
|
347 - 1591 |
1244 |
HSLRVCVDTVR-CNSSTCMMCYK |
0.01052 |
|
347 - 1594 |
1247 |
HSLRVCVDTVR-STCMMCYKRNR |
0.01043 |
|
347 - 1605 |
1258 |
HSLRVCVDTVR-ATRVECTTIVN |
0.03631 |
|
347 - 1627 |
1280 |
HSLRVCVDTVR-GGKGFCKLHNW |
0.0104 |
|
347 - 1634 |
1287 |
HSLRVCVDTVR-LHNWNCVNCDT |
0.01048 |
|
347 - 1637 |
1290 |
HSLRVCVDTVR-WNCVNCDTFCA |
0.01239 |
|
347 - 1641 |
1294 |
HSLRVCVDTVR-NCDTFCAGSTF |
0.99753 |
|
347 - 1731 |
1384 |
HSLRVCVDTVR-DGKSKCEESSA |
0.01101 |
|
347 - 1748 |
1401 |
HSLRVCVDTVR-YSQLMCQPILL |
0.99006 |
|
347 - 1836 |
1489 |
HSLRVCVDTVR-KDVVECLKLSH |
0.01458 |
|
347 - 1852 |
1505 |
HSLRVCVDTVR-VTGDSCNNYML |
0.97945 |
|
347 - 1873 |
1526 |
HSLRVCVDTVR-RDLGACIDCSA |
0.01112 |
|
347 - 1876 |
1529 |
HSLRVCVDTVR-GACIDCSARHI |
0.01107 |
|
347 - 1926 |
1579 |
HSLRVCVDTVR-PFKLTCATTRQ |
0.0104 |
|
417 - 531 |
114 |
KKIKACVEEVT-TVLKKCKSAFY |
0.01114 |
|
417 - 574 |
157 |
KKIKACVEEVT-KLMPVCVETKA |
0.01049 |
|
417 - 856 |
439 |
KKIKACVEEVT-WADNNCYLATA |
0.01162 |
|
417 - 893 |
476 |
KKIKACVEEVT-EAANFCALILA |
0.0104 |
|
417 - 900 |
483 |
KKIKACVEEVT-LILAYCNKTVG |
0.01066 |
|
417 - 926 |
509 |
KKIKACVEEVT-ANLDSCKRVLN |
0.0104 |
|
417 - 934 |
517 |
KKIKACVEEVT-VLNVVCKTCGQ |
0.01077 |
|
417 - 937 |
520 |
KKIKACVEEVT-VVCKTCGQQQT |
0.01041 |
|
417 - 969 |
552 |
KKIKACVEEVT-GVQIPCTCGKQ |
0.01045 |
|
417 - 971 |
554 |
KKIKACVEEVT-QIPCTCGKQAT |
0.01146 |
|
417 - 1005 |
588 |
KKIKACVEEVT-HGTFTCASEYT |
0.01993 |
|
417 - 1015 |
598 |
KKIKACVEEVT-TGNYQCGHYKH |
0.20728 |
|
417 - 1029 |
612 |
KKIKACVEEVT-KETLYCIDGAL |
0.01097 |
|
417 - 1071 |
654 |
KKIKACVEEVT-LDGVVCTEIDP |
0.01302 |
|
417 - 1114 |
697 |
KKIKACVEEVT-NFKFVCDNIKF |
0.02835 |
|
417 - 1191 |
774 |
KKIKACVEEVT-KPNTWCIRCLW |
0.01074 |
|
417 - 1194 |
777 |
KKIKACVEEVT-TWCIRCLWSTK |
0.01091 |
|
417 - 1223 |
806 |
KKIKACVEEVT-MDNLACEDLKP |
0.9675 |
|
417 - 1246 |
829 |
KKIKACVEEVT-KDVLECNVKTT |
0.01053 |
|
417 - 1342 |
925 |
KKIKACVEEVT-NIVTRCLNRVC |
0.01054 |
|
417 - 1347 |
930 |
KKIKACVEEVT-CLNRVCTNYMP |
0.01952 |
|
417 - 1362 |
945 |
KKIKACVEEVT-LLLQLCTFTRS |
0.01827 |
|
417 - 1392 |
975 |
KKIKACVEEVT-SVGKFCLEASF |
0.01097 |
|
417 - 1421 |
1004 |
KKIKACVEEVT-LLLSVCLGSLI |
0.01047 |
|
417 - 1445 |
1028 |
KKIKACVEEVT-GMPSYCTGYRE |
0.01054 |
|
417 - 1464 |
1047 |
KKIKACVEEVT-TIATYCTGSIP |
0.01037 |
|
417 - 1470 |
1053 |
KKIKACVEEVT-TGSIPCSVCLS |
0.19022 |
|
417 - 1473 |
1056 |
KKIKACVEEVT-IPCSVCLSGLD |
0.04124 |
|
417 - 1586 |
1169 |
KKIKACVEEVT-HVVDGCNSSTC |
0.01043 |
|
417 - 1591 |
1174 |
KKIKACVEEVT-CNSSTCMMCYK |
0.01038 |
|
417 - 1594 |
1177 |
KKIKACVEEVT-STCMMCYKRNR |
0.0304 |
|
417 - 1605 |
1188 |
KKIKACVEEVT-ATRVECTTIVN |
0.23862 |
|
417 - 1627 |
1210 |
KKIKACVEEVT-GGKGFCKLHNW |
0.12483 |
|
417 - 1634 |
1217 |
KKIKACVEEVT-LHNWNCVNCDT |
0.01062 |
|
417 - 1637 |
1220 |
KKIKACVEEVT-WNCVNCDTFCA |
0.01189 |
|
417 - 1641 |
1224 |
KKIKACVEEVT-NCDTFCAGSTF |
0.28795 |
|
417 - 1731 |
1314 |
KKIKACVEEVT-DGKSKCEESSA |
0.01113 |
|
417 - 1748 |
1331 |
KKIKACVEEVT-YSQLMCQPILL |
0.01608 |
|
417 - 1836 |
1419 |
KKIKACVEEVT-KDVVECLKLSH |
0.01089 |
|
417 - 1852 |
1435 |
KKIKACVEEVT-VTGDSCNNYML |
0.07189 |
|
417 - 1873 |
1456 |
KKIKACVEEVT-RDLGACIDCSA |
0.01226 |
|
417 - 1876 |
1459 |
KKIKACVEEVT-GACIDCSARHI |
0.0104 |
|
417 - 1926 |
1509 |
KKIKACVEEVT-PFKLTCATTRQ |
0.01039 |
|
531 - 574 |
43 |
TVLKKCKSAFY-KLMPVCVETKA |
0.58597 |
|
531 - 856 |
325 |
TVLKKCKSAFY-WADNNCYLATA |
0.01275 |
|
531 - 893 |
362 |
TVLKKCKSAFY-EAANFCALILA |
0.01424 |
|
531 - 900 |
369 |
TVLKKCKSAFY-LILAYCNKTVG |
0.01888 |
|
531 - 926 |
395 |
TVLKKCKSAFY-ANLDSCKRVLN |
0.01274 |
|
531 - 934 |
403 |
TVLKKCKSAFY-VLNVVCKTCGQ |
0.01407 |
|
531 - 937 |
406 |
TVLKKCKSAFY-VVCKTCGQQQT |
0.01599 |
|
531 - 969 |
438 |
TVLKKCKSAFY-GVQIPCTCGKQ |
0.01216 |
|
531 - 971 |
440 |
TVLKKCKSAFY-QIPCTCGKQAT |
0.70904 |
|
531 - 1005 |
474 |
TVLKKCKSAFY-HGTFTCASEYT |
0.16372 |
|
531 - 1015 |
484 |
TVLKKCKSAFY-TGNYQCGHYKH |
0.95964 |
|
531 - 1029 |
498 |
TVLKKCKSAFY-KETLYCIDGAL |
0.0295 |
|
531 - 1071 |
540 |
TVLKKCKSAFY-LDGVVCTEIDP |
0.02929 |
|
531 - 1114 |
583 |
TVLKKCKSAFY-NFKFVCDNIKF |
0.97039 |
|
531 - 1191 |
660 |
TVLKKCKSAFY-KPNTWCIRCLW |
0.01129 |
|
531 - 1194 |
663 |
TVLKKCKSAFY-TWCIRCLWSTK |
0.12207 |
|
531 - 1223 |
692 |
TVLKKCKSAFY-MDNLACEDLKP |
0.99131 |
|
531 - 1246 |
715 |
TVLKKCKSAFY-KDVLECNVKTT |
0.01414 |
|
531 - 1342 |
811 |
TVLKKCKSAFY-NIVTRCLNRVC |
0.01176 |
|
531 - 1347 |
816 |
TVLKKCKSAFY-CLNRVCTNYMP |
0.40761 |
|
531 - 1362 |
831 |
TVLKKCKSAFY-LLLQLCTFTRS |
0.98164 |
|
531 - 1392 |
861 |
TVLKKCKSAFY-SVGKFCLEASF |
0.01758 |
|
531 - 1421 |
890 |
TVLKKCKSAFY-LLLSVCLGSLI |
0.01279 |
|
531 - 1445 |
914 |
TVLKKCKSAFY-GMPSYCTGYRE |
0.02674 |
|
531 - 1464 |
933 |
TVLKKCKSAFY-TIATYCTGSIP |
0.21097 |
|
531 - 1470 |
939 |
TVLKKCKSAFY-TGSIPCSVCLS |
0.99799 |
|
531 - 1473 |
942 |
TVLKKCKSAFY-IPCSVCLSGLD |
0.64854 |
|
531 - 1586 |
1055 |
TVLKKCKSAFY-HVVDGCNSSTC |
0.01115 |
|
531 - 1591 |
1060 |
TVLKKCKSAFY-CNSSTCMMCYK |
0.01147 |
|
531 - 1594 |
1063 |
TVLKKCKSAFY-STCMMCYKRNR |
0.01311 |
|
531 - 1605 |
1074 |
TVLKKCKSAFY-ATRVECTTIVN |
0.98477 |
|
531 - 1627 |
1096 |
TVLKKCKSAFY-GGKGFCKLHNW |
0.01083 |
|
531 - 1634 |
1103 |
TVLKKCKSAFY-LHNWNCVNCDT |
0.01505 |
|
531 - 1637 |
1106 |
TVLKKCKSAFY-WNCVNCDTFCA |
0.10284 |
|
531 - 1641 |
1110 |
TVLKKCKSAFY-NCDTFCAGSTF |
0.99939 |
|
531 - 1731 |
1200 |
TVLKKCKSAFY-DGKSKCEESSA |
0.02224 |
|
531 - 1748 |
1217 |
TVLKKCKSAFY-YSQLMCQPILL |
0.99934 |
|
531 - 1836 |
1305 |
TVLKKCKSAFY-KDVVECLKLSH |
0.30987 |
|
531 - 1852 |
1321 |
TVLKKCKSAFY-VTGDSCNNYML |
0.99967 |
|
531 - 1873 |
1342 |
TVLKKCKSAFY-RDLGACIDCSA |
0.04013 |
|
531 - 1876 |
1345 |
TVLKKCKSAFY-GACIDCSARHI |
0.01379 |
|
531 - 1926 |
1395 |
TVLKKCKSAFY-PFKLTCATTRQ |
0.01047 |
|
574 - 856 |
282 |
KLMPVCVETKA-WADNNCYLATA |
0.01037 |
|
574 - 893 |
319 |
KLMPVCVETKA-EAANFCALILA |
0.01055 |
|
574 - 900 |
326 |
KLMPVCVETKA-LILAYCNKTVG |
0.01116 |
|
574 - 926 |
352 |
KLMPVCVETKA-ANLDSCKRVLN |
0.01039 |
|
574 - 934 |
360 |
KLMPVCVETKA-VLNVVCKTCGQ |
0.01084 |
|
574 - 937 |
363 |
KLMPVCVETKA-VVCKTCGQQQT |
0.03323 |
|
574 - 969 |
395 |
KLMPVCVETKA-GVQIPCTCGKQ |
0.01037 |
|
574 - 971 |
397 |
KLMPVCVETKA-QIPCTCGKQAT |
0.01039 |
|
574 - 1005 |
431 |
KLMPVCVETKA-HGTFTCASEYT |
0.01051 |
|
574 - 1015 |
441 |
KLMPVCVETKA-TGNYQCGHYKH |
0.01063 |
|
574 - 1029 |
455 |
KLMPVCVETKA-KETLYCIDGAL |
0.01045 |
|
574 - 1071 |
497 |
KLMPVCVETKA-LDGVVCTEIDP |
0.01186 |
|
574 - 1114 |
540 |
KLMPVCVETKA-NFKFVCDNIKF |
0.0161 |
|
574 - 1191 |
617 |
KLMPVCVETKA-KPNTWCIRCLW |
0.01037 |
|
574 - 1194 |
620 |
KLMPVCVETKA-TWCIRCLWSTK |
0.01039 |
|
574 - 1223 |
649 |
KLMPVCVETKA-MDNLACEDLKP |
0.01699 |
|
574 - 1246 |
672 |
KLMPVCVETKA-KDVLECNVKTT |
0.01319 |
|
574 - 1342 |
768 |
KLMPVCVETKA-NIVTRCLNRVC |
0.01039 |
|
574 - 1347 |
773 |
KLMPVCVETKA-CLNRVCTNYMP |
0.01044 |
|
574 - 1362 |
788 |
KLMPVCVETKA-LLLQLCTFTRS |
0.01049 |
|
574 - 1392 |
818 |
KLMPVCVETKA-SVGKFCLEASF |
0.01037 |
|
574 - 1421 |
847 |
KLMPVCVETKA-LLLSVCLGSLI |
0.01037 |
|
574 - 1445 |
871 |
KLMPVCVETKA-GMPSYCTGYRE |
0.01038 |
|
574 - 1464 |
890 |
KLMPVCVETKA-TIATYCTGSIP |
0.01292 |
|
574 - 1470 |
896 |
KLMPVCVETKA-TGSIPCSVCLS |
0.01322 |
|
574 - 1473 |
899 |
KLMPVCVETKA-IPCSVCLSGLD |
0.01115 |
|
574 - 1586 |
1012 |
KLMPVCVETKA-HVVDGCNSSTC |
0.01057 |
|
574 - 1591 |
1017 |
KLMPVCVETKA-CNSSTCMMCYK |
0.01037 |
|
574 - 1594 |
1020 |
KLMPVCVETKA-STCMMCYKRNR |
0.01039 |
|
574 - 1605 |
1031 |
KLMPVCVETKA-ATRVECTTIVN |
0.01039 |
|
574 - 1627 |
1053 |
KLMPVCVETKA-GGKGFCKLHNW |
0.01038 |
|
574 - 1634 |
1060 |
KLMPVCVETKA-LHNWNCVNCDT |
0.01039 |
|
574 - 1637 |
1063 |
KLMPVCVETKA-WNCVNCDTFCA |
0.01137 |
|
574 - 1641 |
1067 |
KLMPVCVETKA-NCDTFCAGSTF |
0.02021 |
|
574 - 1731 |
1157 |
KLMPVCVETKA-DGKSKCEESSA |
0.01044 |
|
574 - 1748 |
1174 |
KLMPVCVETKA-YSQLMCQPILL |
0.01385 |
|
574 - 1836 |
1262 |
KLMPVCVETKA-KDVVECLKLSH |
0.01044 |
|
574 - 1852 |
1278 |
KLMPVCVETKA-VTGDSCNNYML |
0.02149 |
|
574 - 1873 |
1299 |
KLMPVCVETKA-RDLGACIDCSA |
0.01038 |
|
574 - 1876 |
1302 |
KLMPVCVETKA-GACIDCSARHI |
0.01197 |
|
574 - 1926 |
1352 |
KLMPVCVETKA-PFKLTCATTRQ |
0.0104 |
|
856 - 893 |
37 |
WADNNCYLATA-EAANFCALILA |
0.01042 |
|
856 - 900 |
44 |
WADNNCYLATA-LILAYCNKTVG |
0.0106 |
|
856 - 926 |
70 |
WADNNCYLATA-ANLDSCKRVLN |
0.01037 |
|
856 - 934 |
78 |
WADNNCYLATA-VLNVVCKTCGQ |
0.01037 |
|
856 - 937 |
81 |
WADNNCYLATA-VVCKTCGQQQT |
0.02246 |
|
856 - 969 |
113 |
WADNNCYLATA-GVQIPCTCGKQ |
0.01037 |
|
856 - 971 |
115 |
WADNNCYLATA-QIPCTCGKQAT |
0.01037 |
|
856 - 1005 |
149 |
WADNNCYLATA-HGTFTCASEYT |
0.01037 |
|
856 - 1015 |
159 |
WADNNCYLATA-TGNYQCGHYKH |
0.01037 |
|
856 - 1029 |
173 |
WADNNCYLATA-KETLYCIDGAL |
0.01038 |
|
856 - 1071 |
215 |
WADNNCYLATA-LDGVVCTEIDP |
0.0104 |
|
856 - 1114 |
258 |
WADNNCYLATA-NFKFVCDNIKF |
0.01061 |
|
856 - 1191 |
335 |
WADNNCYLATA-KPNTWCIRCLW |
0.01037 |
|
856 - 1194 |
338 |
WADNNCYLATA-TWCIRCLWSTK |
0.01038 |
|
856 - 1223 |
367 |
WADNNCYLATA-MDNLACEDLKP |
0.01039 |
|
856 - 1246 |
390 |
WADNNCYLATA-KDVLECNVKTT |
0.01045 |
|
856 - 1342 |
486 |
WADNNCYLATA-NIVTRCLNRVC |
0.01038 |
|
856 - 1347 |
491 |
WADNNCYLATA-CLNRVCTNYMP |
0.01037 |
|
856 - 1362 |
506 |
WADNNCYLATA-LLLQLCTFTRS |
0.01045 |
|
856 - 1392 |
536 |
WADNNCYLATA-SVGKFCLEASF |
0.01037 |
|
856 - 1421 |
565 |
WADNNCYLATA-LLLSVCLGSLI |
0.01037 |
|
856 - 1445 |
589 |
WADNNCYLATA-GMPSYCTGYRE |
0.01037 |
|
856 - 1464 |
608 |
WADNNCYLATA-TIATYCTGSIP |
0.01415 |
|
856 - 1470 |
614 |
WADNNCYLATA-TGSIPCSVCLS |
0.01049 |
|
856 - 1473 |
617 |
WADNNCYLATA-IPCSVCLSGLD |
0.01047 |
|
856 - 1586 |
730 |
WADNNCYLATA-HVVDGCNSSTC |
0.0104 |
|
856 - 1591 |
735 |
WADNNCYLATA-CNSSTCMMCYK |
0.01037 |
|
856 - 1594 |
738 |
WADNNCYLATA-STCMMCYKRNR |
0.01037 |
|
856 - 1605 |
749 |
WADNNCYLATA-ATRVECTTIVN |
0.01037 |
|
856 - 1627 |
771 |
WADNNCYLATA-GGKGFCKLHNW |
0.0104 |
|
856 - 1634 |
778 |
WADNNCYLATA-LHNWNCVNCDT |
0.01037 |
|
856 - 1637 |
781 |
WADNNCYLATA-WNCVNCDTFCA |
0.01097 |
|
856 - 1641 |
785 |
WADNNCYLATA-NCDTFCAGSTF |
0.97618 |
|
856 - 1731 |
875 |
WADNNCYLATA-DGKSKCEESSA |
0.01037 |
|
856 - 1748 |
892 |
WADNNCYLATA-YSQLMCQPILL |
0.01134 |
|
856 - 1836 |
980 |
WADNNCYLATA-KDVVECLKLSH |
0.01037 |
|
856 - 1852 |
996 |
WADNNCYLATA-VTGDSCNNYML |
0.0106 |
|
856 - 1873 |
1017 |
WADNNCYLATA-RDLGACIDCSA |
0.01037 |
|
856 - 1876 |
1020 |
WADNNCYLATA-GACIDCSARHI |
0.011 |
|
856 - 1926 |
1070 |
WADNNCYLATA-PFKLTCATTRQ |
0.01037 |
|
893 - 900 |
7 |
EAANFCALILA-LILAYCNKTVG |
0.01043 |
|
893 - 926 |
33 |
EAANFCALILA-ANLDSCKRVLN |
0.01068 |
|
893 - 934 |
41 |
EAANFCALILA-VLNVVCKTCGQ |
0.01127 |
|
893 - 937 |
44 |
EAANFCALILA-VVCKTCGQQQT |
0.01058 |
|
893 - 969 |
76 |
EAANFCALILA-GVQIPCTCGKQ |
0.01041 |
|
893 - 971 |
78 |
EAANFCALILA-QIPCTCGKQAT |
0.01062 |
|
893 - 1005 |
112 |
EAANFCALILA-HGTFTCASEYT |
0.01789 |
|
893 - 1015 |
122 |
EAANFCALILA-TGNYQCGHYKH |
0.04748 |
|
893 - 1029 |
136 |
EAANFCALILA-KETLYCIDGAL |
0.01063 |
|
893 - 1071 |
178 |
EAANFCALILA-LDGVVCTEIDP |
0.01781 |
|
893 - 1114 |
221 |
EAANFCALILA-NFKFVCDNIKF |
0.23477 |
|
893 - 1191 |
298 |
EAANFCALILA-KPNTWCIRCLW |
0.01075 |
|
893 - 1194 |
301 |
EAANFCALILA-TWCIRCLWSTK |
0.0104 |
|
893 - 1223 |
330 |
EAANFCALILA-MDNLACEDLKP |
0.01141 |
|
893 - 1246 |
353 |
EAANFCALILA-KDVLECNVKTT |
0.01048 |
|
893 - 1342 |
449 |
EAANFCALILA-NIVTRCLNRVC |
0.01037 |
|
893 - 1347 |
454 |
EAANFCALILA-CLNRVCTNYMP |
0.01066 |
|
893 - 1362 |
469 |
EAANFCALILA-LLLQLCTFTRS |
0.01043 |
|
893 - 1392 |
499 |
EAANFCALILA-SVGKFCLEASF |
0.01038 |
|
893 - 1421 |
528 |
EAANFCALILA-LLLSVCLGSLI |
0.01037 |
|
893 - 1445 |
552 |
EAANFCALILA-GMPSYCTGYRE |
0.0105 |
|
893 - 1464 |
571 |
EAANFCALILA-TIATYCTGSIP |
0.0105 |
|
893 - 1470 |
577 |
EAANFCALILA-TGSIPCSVCLS |
0.11883 |
|
893 - 1473 |
580 |
EAANFCALILA-IPCSVCLSGLD |
0.01133 |
|
893 - 1586 |
693 |
EAANFCALILA-HVVDGCNSSTC |
0.01044 |
|
893 - 1591 |
698 |
EAANFCALILA-CNSSTCMMCYK |
0.01039 |
|
893 - 1594 |
701 |
EAANFCALILA-STCMMCYKRNR |
0.0117 |
|
893 - 1605 |
712 |
EAANFCALILA-ATRVECTTIVN |
0.01048 |
|
893 - 1627 |
734 |
EAANFCALILA-GGKGFCKLHNW |
0.20434 |
|
893 - 1634 |
741 |
EAANFCALILA-LHNWNCVNCDT |
0.01209 |
|
893 - 1637 |
744 |
EAANFCALILA-WNCVNCDTFCA |
0.01346 |
|
893 - 1641 |
748 |
EAANFCALILA-NCDTFCAGSTF |
0.39424 |
|
893 - 1731 |
838 |
EAANFCALILA-DGKSKCEESSA |
0.01086 |
|
893 - 1748 |
855 |
EAANFCALILA-YSQLMCQPILL |
0.01615 |
|
893 - 1836 |
943 |
EAANFCALILA-KDVVECLKLSH |
0.01039 |
|
893 - 1852 |
959 |
EAANFCALILA-VTGDSCNNYML |
0.76236 |
|
893 - 1873 |
980 |
EAANFCALILA-RDLGACIDCSA |
0.01039 |
|
893 - 1876 |
983 |
EAANFCALILA-GACIDCSARHI |
0.01043 |
|
893 - 1926 |
1033 |
EAANFCALILA-PFKLTCATTRQ |
0.01039 |
|
900 - 926 |
26 |
LILAYCNKTVG-ANLDSCKRVLN |
0.01044 |
|
900 - 934 |
34 |
LILAYCNKTVG-VLNVVCKTCGQ |
0.0137 |
|
900 - 937 |
37 |
LILAYCNKTVG-VVCKTCGQQQT |
0.01086 |
|
900 - 969 |
69 |
LILAYCNKTVG-GVQIPCTCGKQ |
0.01048 |
|
900 - 971 |
71 |
LILAYCNKTVG-QIPCTCGKQAT |
0.06193 |
|
900 - 1005 |
105 |
LILAYCNKTVG-HGTFTCASEYT |
0.22033 |
|
900 - 1015 |
115 |
LILAYCNKTVG-TGNYQCGHYKH |
0.03711 |
|
900 - 1029 |
129 |
LILAYCNKTVG-KETLYCIDGAL |
0.01091 |
|
900 - 1071 |
171 |
LILAYCNKTVG-LDGVVCTEIDP |
0.08216 |
|
900 - 1114 |
214 |
LILAYCNKTVG-NFKFVCDNIKF |
0.96887 |
|
900 - 1191 |
291 |
LILAYCNKTVG-KPNTWCIRCLW |
0.01217 |
|
900 - 1194 |
294 |
LILAYCNKTVG-TWCIRCLWSTK |
0.01358 |
|
900 - 1223 |
323 |
LILAYCNKTVG-MDNLACEDLKP |
0.99663 |
|
900 - 1246 |
346 |
LILAYCNKTVG-KDVLECNVKTT |
0.01116 |
|
900 - 1342 |
442 |
LILAYCNKTVG-NIVTRCLNRVC |
0.01088 |
|
900 - 1347 |
447 |
LILAYCNKTVG-CLNRVCTNYMP |
0.98893 |
|
900 - 1362 |
462 |
LILAYCNKTVG-LLLQLCTFTRS |
0.99448 |
|
900 - 1392 |
492 |
LILAYCNKTVG-SVGKFCLEASF |
0.03342 |
|
900 - 1421 |
521 |
LILAYCNKTVG-LLLSVCLGSLI |
0.01239 |
|
900 - 1445 |
545 |
LILAYCNKTVG-GMPSYCTGYRE |
0.01094 |
|
900 - 1464 |
564 |
LILAYCNKTVG-TIATYCTGSIP |
0.01138 |
|
900 - 1470 |
570 |
LILAYCNKTVG-TGSIPCSVCLS |
0.76296 |
|
900 - 1473 |
573 |
LILAYCNKTVG-IPCSVCLSGLD |
0.01548 |
|
900 - 1586 |
686 |
LILAYCNKTVG-HVVDGCNSSTC |
0.01072 |
|
900 - 1591 |
691 |
LILAYCNKTVG-CNSSTCMMCYK |
0.01046 |
|
900 - 1594 |
694 |
LILAYCNKTVG-STCMMCYKRNR |
0.01065 |
|
900 - 1605 |
705 |
LILAYCNKTVG-ATRVECTTIVN |
0.80037 |
|
900 - 1627 |
727 |
LILAYCNKTVG-GGKGFCKLHNW |
0.01147 |
|
900 - 1634 |
734 |
LILAYCNKTVG-LHNWNCVNCDT |
0.02149 |
|
900 - 1637 |
737 |
LILAYCNKTVG-WNCVNCDTFCA |
0.01127 |
|
900 - 1641 |
741 |
LILAYCNKTVG-NCDTFCAGSTF |
0.05783 |
|
900 - 1731 |
831 |
LILAYCNKTVG-DGKSKCEESSA |
0.03303 |
|
900 - 1748 |
848 |
LILAYCNKTVG-YSQLMCQPILL |
0.36466 |
|
900 - 1836 |
936 |
LILAYCNKTVG-KDVVECLKLSH |
0.01354 |
|
900 - 1852 |
952 |
LILAYCNKTVG-VTGDSCNNYML |
0.15745 |
|
900 - 1873 |
973 |
LILAYCNKTVG-RDLGACIDCSA |
0.88205 |
|
900 - 1876 |
976 |
LILAYCNKTVG-GACIDCSARHI |
0.01055 |
|
900 - 1926 |
1026 |
LILAYCNKTVG-PFKLTCATTRQ |
0.01093 |
|
926 - 934 |
8 |
ANLDSCKRVLN-VLNVVCKTCGQ |
0.01037 |
|
926 - 937 |
11 |
ANLDSCKRVLN-VVCKTCGQQQT |
0.99821 |
|
926 - 969 |
43 |
ANLDSCKRVLN-GVQIPCTCGKQ |
0.01064 |
|
926 - 971 |
45 |
ANLDSCKRVLN-QIPCTCGKQAT |
0.01042 |
|
926 - 1005 |
79 |
ANLDSCKRVLN-HGTFTCASEYT |
0.01272 |
|
926 - 1015 |
89 |
ANLDSCKRVLN-TGNYQCGHYKH |
0.02159 |
|
926 - 1029 |
103 |
ANLDSCKRVLN-KETLYCIDGAL |
0.01897 |
|
926 - 1071 |
145 |
ANLDSCKRVLN-LDGVVCTEIDP |
0.01041 |
|
926 - 1114 |
188 |
ANLDSCKRVLN-NFKFVCDNIKF |
0.0104 |
|
926 - 1191 |
265 |
ANLDSCKRVLN-KPNTWCIRCLW |
0.01131 |
|
926 - 1194 |
268 |
ANLDSCKRVLN-TWCIRCLWSTK |
0.99882 |
|
926 - 1223 |
297 |
ANLDSCKRVLN-MDNLACEDLKP |
0.07596 |
|
926 - 1246 |
320 |
ANLDSCKRVLN-KDVLECNVKTT |
0.99796 |
|
926 - 1342 |
416 |
ANLDSCKRVLN-NIVTRCLNRVC |
0.01114 |
|
926 - 1347 |
421 |
ANLDSCKRVLN-CLNRVCTNYMP |
0.0104 |
|
926 - 1362 |
436 |
ANLDSCKRVLN-LLLQLCTFTRS |
0.01266 |
|
926 - 1392 |
466 |
ANLDSCKRVLN-SVGKFCLEASF |
0.01052 |
|
926 - 1421 |
495 |
ANLDSCKRVLN-LLLSVCLGSLI |
0.01041 |
|
926 - 1445 |
519 |
ANLDSCKRVLN-GMPSYCTGYRE |
0.01042 |
|
926 - 1464 |
538 |
ANLDSCKRVLN-TIATYCTGSIP |
0.99956 |
|
926 - 1470 |
544 |
ANLDSCKRVLN-TGSIPCSVCLS |
0.01037 |
|
926 - 1473 |
547 |
ANLDSCKRVLN-IPCSVCLSGLD |
0.03663 |
|
926 - 1586 |
660 |
ANLDSCKRVLN-HVVDGCNSSTC |
0.67392 |
|
926 - 1591 |
665 |
ANLDSCKRVLN-CNSSTCMMCYK |
0.01037 |
|
926 - 1594 |
668 |
ANLDSCKRVLN-STCMMCYKRNR |
0.69395 |
|
926 - 1605 |
679 |
ANLDSCKRVLN-ATRVECTTIVN |
0.01235 |
|
926 - 1627 |
701 |
ANLDSCKRVLN-GGKGFCKLHNW |
0.04492 |
|
926 - 1634 |
708 |
ANLDSCKRVLN-LHNWNCVNCDT |
0.99803 |
|
926 - 1637 |
711 |
ANLDSCKRVLN-WNCVNCDTFCA |
0.97965 |
|
926 - 1641 |
715 |
ANLDSCKRVLN-NCDTFCAGSTF |
0.01131 |
|
926 - 1731 |
805 |
ANLDSCKRVLN-DGKSKCEESSA |
0.01037 |
|
926 - 1748 |
822 |
ANLDSCKRVLN-YSQLMCQPILL |
0.01037 |
|
926 - 1836 |
910 |
ANLDSCKRVLN-KDVVECLKLSH |
0.01042 |
|
926 - 1852 |
926 |
ANLDSCKRVLN-VTGDSCNNYML |
0.01083 |
|
926 - 1873 |
947 |
ANLDSCKRVLN-RDLGACIDCSA |
0.01246 |
|
926 - 1876 |
950 |
ANLDSCKRVLN-GACIDCSARHI |
0.998 |
|
926 - 1926 |
1000 |
ANLDSCKRVLN-PFKLTCATTRQ |
0.01091 |
|
934 - 937 |
3 |
VLNVVCKTCGQ-VVCKTCGQQQT |
0.0114 |
|
934 - 969 |
35 |
VLNVVCKTCGQ-GVQIPCTCGKQ |
0.07884 |
|
934 - 971 |
37 |
VLNVVCKTCGQ-QIPCTCGKQAT |
0.01042 |
|
934 - 1005 |
71 |
VLNVVCKTCGQ-HGTFTCASEYT |
0.16653 |
|
934 - 1015 |
81 |
VLNVVCKTCGQ-TGNYQCGHYKH |
0.98999 |
|
934 - 1029 |
95 |
VLNVVCKTCGQ-KETLYCIDGAL |
0.01091 |
|
934 - 1071 |
137 |
VLNVVCKTCGQ-LDGVVCTEIDP |
0.99811 |
|
934 - 1114 |
180 |
VLNVVCKTCGQ-NFKFVCDNIKF |
0.01347 |
|
934 - 1191 |
257 |
VLNVVCKTCGQ-KPNTWCIRCLW |
0.05673 |
|
934 - 1194 |
260 |
VLNVVCKTCGQ-TWCIRCLWSTK |
0.01123 |
|
934 - 1223 |
289 |
VLNVVCKTCGQ-MDNLACEDLKP |
0.99771 |
|
934 - 1246 |
312 |
VLNVVCKTCGQ-KDVLECNVKTT |
0.01696 |
|
934 - 1342 |
408 |
VLNVVCKTCGQ-NIVTRCLNRVC |
0.01064 |
|
934 - 1347 |
413 |
VLNVVCKTCGQ-CLNRVCTNYMP |
0.38734 |
|
934 - 1362 |
428 |
VLNVVCKTCGQ-LLLQLCTFTRS |
0.99335 |
|
934 - 1392 |
458 |
VLNVVCKTCGQ-SVGKFCLEASF |
0.01105 |
|
934 - 1421 |
487 |
VLNVVCKTCGQ-LLLSVCLGSLI |
0.0108 |
|
934 - 1445 |
511 |
VLNVVCKTCGQ-GMPSYCTGYRE |
0.3438 |
|
934 - 1464 |
530 |
VLNVVCKTCGQ-TIATYCTGSIP |
0.02894 |
|
934 - 1470 |
536 |
VLNVVCKTCGQ-TGSIPCSVCLS |
0.01143 |
|
934 - 1473 |
539 |
VLNVVCKTCGQ-IPCSVCLSGLD |
0.01496 |
|
934 - 1586 |
652 |
VLNVVCKTCGQ-HVVDGCNSSTC |
0.01038 |
|
934 - 1591 |
657 |
VLNVVCKTCGQ-CNSSTCMMCYK |
0.0104 |
|
934 - 1594 |
660 |
VLNVVCKTCGQ-STCMMCYKRNR |
0.8515 |
|
934 - 1605 |
671 |
VLNVVCKTCGQ-ATRVECTTIVN |
0.99544 |
|
934 - 1627 |
693 |
VLNVVCKTCGQ-GGKGFCKLHNW |
0.99733 |
|
934 - 1634 |
700 |
VLNVVCKTCGQ-LHNWNCVNCDT |
0.30542 |
|
934 - 1637 |
703 |
VLNVVCKTCGQ-WNCVNCDTFCA |
0.03455 |
|
934 - 1641 |
707 |
VLNVVCKTCGQ-NCDTFCAGSTF |
0.99845 |
|
934 - 1731 |
797 |
VLNVVCKTCGQ-DGKSKCEESSA |
0.03308 |
|
934 - 1748 |
814 |
VLNVVCKTCGQ-YSQLMCQPILL |
0.01188 |
|
934 - 1836 |
902 |
VLNVVCKTCGQ-KDVVECLKLSH |
0.01072 |
|
934 - 1852 |
918 |
VLNVVCKTCGQ-VTGDSCNNYML |
0.99536 |
|
934 - 1873 |
939 |
VLNVVCKTCGQ-RDLGACIDCSA |
0.96893 |
|
934 - 1876 |
942 |
VLNVVCKTCGQ-GACIDCSARHI |
0.0414 |
|
934 - 1926 |
992 |
VLNVVCKTCGQ-PFKLTCATTRQ |
0.01103 |
|
937 - 969 |
32 |
VVCKTCGQQQT-GVQIPCTCGKQ |
0.01042 |
|
937 - 971 |
34 |
VVCKTCGQQQT-QIPCTCGKQAT |
0.01182 |
|
937 - 1005 |
68 |
VVCKTCGQQQT-HGTFTCASEYT |
0.0109 |
|
937 - 1015 |
78 |
VVCKTCGQQQT-TGNYQCGHYKH |
0.01235 |
|
937 - 1029 |
92 |
VVCKTCGQQQT-KETLYCIDGAL |
0.01172 |
|
937 - 1071 |
134 |
VVCKTCGQQQT-LDGVVCTEIDP |
0.01052 |
|
937 - 1114 |
177 |
VVCKTCGQQQT-NFKFVCDNIKF |
0.01551 |
|
937 - 1191 |
254 |
VVCKTCGQQQT-KPNTWCIRCLW |
0.01041 |
|
937 - 1194 |
257 |
VVCKTCGQQQT-TWCIRCLWSTK |
0.01042 |
|
937 - 1223 |
286 |
VVCKTCGQQQT-MDNLACEDLKP |
0.01047 |
|
937 - 1246 |
309 |
VVCKTCGQQQT-KDVLECNVKTT |
0.01062 |
|
937 - 1342 |
405 |
VVCKTCGQQQT-NIVTRCLNRVC |
0.01037 |
|
937 - 1347 |
410 |
VVCKTCGQQQT-CLNRVCTNYMP |
0.01037 |
|
937 - 1362 |
425 |
VVCKTCGQQQT-LLLQLCTFTRS |
0.01044 |
|
937 - 1392 |
455 |
VVCKTCGQQQT-SVGKFCLEASF |
0.01037 |
|
937 - 1421 |
484 |
VVCKTCGQQQT-LLLSVCLGSLI |
0.01038 |
|
937 - 1445 |
508 |
VVCKTCGQQQT-GMPSYCTGYRE |
0.01043 |
|
937 - 1464 |
527 |
VVCKTCGQQQT-TIATYCTGSIP |
0.01823 |
|
937 - 1470 |
533 |
VVCKTCGQQQT-TGSIPCSVCLS |
0.02492 |
|
937 - 1473 |
536 |
VVCKTCGQQQT-IPCSVCLSGLD |
0.01061 |
|
937 - 1586 |
649 |
VVCKTCGQQQT-HVVDGCNSSTC |
0.01056 |
|
937 - 1591 |
654 |
VVCKTCGQQQT-CNSSTCMMCYK |
0.01039 |
|
937 - 1594 |
657 |
VVCKTCGQQQT-STCMMCYKRNR |
0.01042 |
|
937 - 1605 |
668 |
VVCKTCGQQQT-ATRVECTTIVN |
0.01037 |
|
937 - 1627 |
690 |
VVCKTCGQQQT-GGKGFCKLHNW |
0.02857 |
|
937 - 1634 |
697 |
VVCKTCGQQQT-LHNWNCVNCDT |
0.01045 |
|
937 - 1637 |
700 |
VVCKTCGQQQT-WNCVNCDTFCA |
0.01085 |
|
937 - 1641 |
704 |
VVCKTCGQQQT-NCDTFCAGSTF |
0.99993 |
|
937 - 1731 |
794 |
VVCKTCGQQQT-DGKSKCEESSA |
0.01042 |
|
937 - 1748 |
811 |
VVCKTCGQQQT-YSQLMCQPILL |
0.03076 |
|
937 - 1836 |
899 |
VVCKTCGQQQT-KDVVECLKLSH |
0.01078 |
|
937 - 1852 |
915 |
VVCKTCGQQQT-VTGDSCNNYML |
0.0378 |
|
937 - 1873 |
936 |
VVCKTCGQQQT-RDLGACIDCSA |
0.01037 |
|
937 - 1876 |
939 |
VVCKTCGQQQT-GACIDCSARHI |
0.01046 |
|
937 - 1926 |
989 |
VVCKTCGQQQT-PFKLTCATTRQ |
0.01046 |
|
969 - 971 |
2 |
GVQIPCTCGKQ-QIPCTCGKQAT |
0.01038 |
|
969 - 1005 |
36 |
GVQIPCTCGKQ-HGTFTCASEYT |
0.01037 |
|
969 - 1015 |
46 |
GVQIPCTCGKQ-TGNYQCGHYKH |
0.07692 |
|
969 - 1029 |
60 |
GVQIPCTCGKQ-KETLYCIDGAL |
0.01037 |
|
969 - 1071 |
102 |
GVQIPCTCGKQ-LDGVVCTEIDP |
0.01038 |
|
969 - 1114 |
145 |
GVQIPCTCGKQ-NFKFVCDNIKF |
0.01037 |
|
969 - 1191 |
222 |
GVQIPCTCGKQ-KPNTWCIRCLW |
0.01041 |
|
969 - 1194 |
225 |
GVQIPCTCGKQ-TWCIRCLWSTK |
0.0104 |
|
969 - 1223 |
254 |
GVQIPCTCGKQ-MDNLACEDLKP |
0.01043 |
|
969 - 1246 |
277 |
GVQIPCTCGKQ-KDVLECNVKTT |
0.01037 |
|
969 - 1342 |
373 |
GVQIPCTCGKQ-NIVTRCLNRVC |
0.01039 |
|
969 - 1347 |
378 |
GVQIPCTCGKQ-CLNRVCTNYMP |
0.01037 |
|
969 - 1362 |
393 |
GVQIPCTCGKQ-LLLQLCTFTRS |
0.01125 |
|
969 - 1392 |
423 |
GVQIPCTCGKQ-SVGKFCLEASF |
0.01159 |
|
969 - 1421 |
452 |
GVQIPCTCGKQ-LLLSVCLGSLI |
0.78375 |
|
969 - 1445 |
476 |
GVQIPCTCGKQ-GMPSYCTGYRE |
0.01117 |
|
969 - 1464 |
495 |
GVQIPCTCGKQ-TIATYCTGSIP |
0.0104 |
|
969 - 1470 |
501 |
GVQIPCTCGKQ-TGSIPCSVCLS |
0.01041 |
|
969 - 1473 |
504 |
GVQIPCTCGKQ-IPCSVCLSGLD |
0.01037 |
|
969 - 1586 |
617 |
GVQIPCTCGKQ-HVVDGCNSSTC |
0.01037 |
|
969 - 1591 |
622 |
GVQIPCTCGKQ-CNSSTCMMCYK |
0.01064 |
|
969 - 1594 |
625 |
GVQIPCTCGKQ-STCMMCYKRNR |
0.01599 |
|
969 - 1605 |
636 |
GVQIPCTCGKQ-ATRVECTTIVN |
0.01037 |
|
969 - 1627 |
658 |
GVQIPCTCGKQ-GGKGFCKLHNW |
0.50443 |
|
969 - 1634 |
665 |
GVQIPCTCGKQ-LHNWNCVNCDT |
0.01037 |
|
969 - 1637 |
668 |
GVQIPCTCGKQ-WNCVNCDTFCA |
0.0104 |
|
969 - 1641 |
672 |
GVQIPCTCGKQ-NCDTFCAGSTF |
0.72664 |
|
969 - 1731 |
762 |
GVQIPCTCGKQ-DGKSKCEESSA |
0.01039 |
|
969 - 1748 |
779 |
GVQIPCTCGKQ-YSQLMCQPILL |
0.01043 |
|
969 - 1836 |
867 |
GVQIPCTCGKQ-KDVVECLKLSH |
0.01037 |
|
969 - 1852 |
883 |
GVQIPCTCGKQ-VTGDSCNNYML |
0.01039 |
|
969 - 1873 |
904 |
GVQIPCTCGKQ-RDLGACIDCSA |
0.03494 |
|
969 - 1876 |
907 |
GVQIPCTCGKQ-GACIDCSARHI |
0.01044 |
|
969 - 1926 |
957 |
GVQIPCTCGKQ-PFKLTCATTRQ |
0.01037 |
|
971 - 1005 |
34 |
QIPCTCGKQAT-HGTFTCASEYT |
0.01044 |
|
971 - 1015 |
44 |
QIPCTCGKQAT-TGNYQCGHYKH |
0.01042 |
|
971 - 1029 |
58 |
QIPCTCGKQAT-KETLYCIDGAL |
0.01038 |
|
971 - 1071 |
100 |
QIPCTCGKQAT-LDGVVCTEIDP |
0.01041 |
|
971 - 1114 |
143 |
QIPCTCGKQAT-NFKFVCDNIKF |
0.01044 |
|
971 - 1191 |
220 |
QIPCTCGKQAT-KPNTWCIRCLW |
0.01037 |
|
971 - 1194 |
223 |
QIPCTCGKQAT-TWCIRCLWSTK |
0.0104 |
|
971 - 1223 |
252 |
QIPCTCGKQAT-MDNLACEDLKP |
0.01483 |
|
971 - 1246 |
275 |
QIPCTCGKQAT-KDVLECNVKTT |
0.0104 |
|
971 - 1342 |
371 |
QIPCTCGKQAT-NIVTRCLNRVC |
0.01037 |
|
971 - 1347 |
376 |
QIPCTCGKQAT-CLNRVCTNYMP |
0.01043 |
|
971 - 1362 |
391 |
QIPCTCGKQAT-LLLQLCTFTRS |
0.01063 |
|
971 - 1392 |
421 |
QIPCTCGKQAT-SVGKFCLEASF |
0.01037 |
|
971 - 1421 |
450 |
QIPCTCGKQAT-LLLSVCLGSLI |
0.01047 |
|
971 - 1445 |
474 |
QIPCTCGKQAT-GMPSYCTGYRE |
0.01037 |
|
971 - 1464 |
493 |
QIPCTCGKQAT-TIATYCTGSIP |
0.01126 |
|
971 - 1470 |
499 |
QIPCTCGKQAT-TGSIPCSVCLS |
0.0119 |
|
971 - 1473 |
502 |
QIPCTCGKQAT-IPCSVCLSGLD |
0.01058 |
|
971 - 1586 |
615 |
QIPCTCGKQAT-HVVDGCNSSTC |
0.01038 |
|
971 - 1591 |
620 |
QIPCTCGKQAT-CNSSTCMMCYK |
0.01037 |
|
971 - 1594 |
623 |
QIPCTCGKQAT-STCMMCYKRNR |
0.01041 |
|
971 - 1605 |
634 |
QIPCTCGKQAT-ATRVECTTIVN |
0.0106 |
|
971 - 1627 |
656 |
QIPCTCGKQAT-GGKGFCKLHNW |
0.01049 |
|
971 - 1634 |
663 |
QIPCTCGKQAT-LHNWNCVNCDT |
0.01038 |
|
971 - 1637 |
666 |
QIPCTCGKQAT-WNCVNCDTFCA |
0.01051 |
|
971 - 1641 |
670 |
QIPCTCGKQAT-NCDTFCAGSTF |
0.02113 |
|
971 - 1731 |
760 |
QIPCTCGKQAT-DGKSKCEESSA |
0.01037 |
|
971 - 1748 |
777 |
QIPCTCGKQAT-YSQLMCQPILL |
0.0104 |
|
971 - 1836 |
865 |
QIPCTCGKQAT-KDVVECLKLSH |
0.01037 |
|
971 - 1852 |
881 |
QIPCTCGKQAT-VTGDSCNNYML |
0.01069 |
|
971 - 1873 |
902 |
QIPCTCGKQAT-RDLGACIDCSA |
0.01111 |
|
971 - 1876 |
905 |
QIPCTCGKQAT-GACIDCSARHI |
0.01044 |
|
971 - 1926 |
955 |
QIPCTCGKQAT-PFKLTCATTRQ |
0.01037 |
|
1005 - 1015 |
10 |
HGTFTCASEYT-TGNYQCGHYKH |
0.97507 |
|
1005 - 1029 |
24 |
HGTFTCASEYT-KETLYCIDGAL |
0.01038 |
|
1005 - 1071 |
66 |
HGTFTCASEYT-LDGVVCTEIDP |
0.96444 |
|
1005 - 1114 |
109 |
HGTFTCASEYT-NFKFVCDNIKF |
0.01106 |
|
1005 - 1191 |
186 |
HGTFTCASEYT-KPNTWCIRCLW |
0.32284 |
|
1005 - 1194 |
189 |
HGTFTCASEYT-TWCIRCLWSTK |
0.01042 |
|
1005 - 1223 |
218 |
HGTFTCASEYT-MDNLACEDLKP |
0.01037 |
|
1005 - 1246 |
241 |
HGTFTCASEYT-KDVLECNVKTT |
0.01054 |
|
1005 - 1342 |
337 |
HGTFTCASEYT-NIVTRCLNRVC |
0.01038 |
|
1005 - 1347 |
342 |
HGTFTCASEYT-CLNRVCTNYMP |
0.0104 |
|
1005 - 1362 |
357 |
HGTFTCASEYT-LLLQLCTFTRS |
0.01045 |
|
1005 - 1392 |
387 |
HGTFTCASEYT-SVGKFCLEASF |
0.0104 |
|
1005 - 1421 |
416 |
HGTFTCASEYT-LLLSVCLGSLI |
0.01037 |
|
1005 - 1445 |
440 |
HGTFTCASEYT-GMPSYCTGYRE |
0.01698 |
|
1005 - 1464 |
459 |
HGTFTCASEYT-TIATYCTGSIP |
0.01098 |
|
1005 - 1470 |
465 |
HGTFTCASEYT-TGSIPCSVCLS |
0.01055 |
|
1005 - 1473 |
468 |
HGTFTCASEYT-IPCSVCLSGLD |
0.01038 |
|
1005 - 1586 |
581 |
HGTFTCASEYT-HVVDGCNSSTC |
0.01039 |
|
1005 - 1591 |
586 |
HGTFTCASEYT-CNSSTCMMCYK |
0.01051 |
|
1005 - 1594 |
589 |
HGTFTCASEYT-STCMMCYKRNR |
0.03833 |
|
1005 - 1605 |
600 |
HGTFTCASEYT-ATRVECTTIVN |
0.01101 |
|
1005 - 1627 |
622 |
HGTFTCASEYT-GGKGFCKLHNW |
0.99284 |
|
1005 - 1634 |
629 |
HGTFTCASEYT-LHNWNCVNCDT |
0.01074 |
|
1005 - 1637 |
632 |
HGTFTCASEYT-WNCVNCDTFCA |
0.01054 |
|
1005 - 1641 |
636 |
HGTFTCASEYT-NCDTFCAGSTF |
0.01693 |
|
1005 - 1731 |
726 |
HGTFTCASEYT-DGKSKCEESSA |
0.02559 |
|
1005 - 1748 |
743 |
HGTFTCASEYT-YSQLMCQPILL |
0.01122 |
|
1005 - 1836 |
831 |
HGTFTCASEYT-KDVVECLKLSH |
0.01038 |
|
1005 - 1852 |
847 |
HGTFTCASEYT-VTGDSCNNYML |
0.04967 |
|
1005 - 1873 |
868 |
HGTFTCASEYT-RDLGACIDCSA |
0.01037 |
|
1005 - 1876 |
871 |
HGTFTCASEYT-GACIDCSARHI |
0.79428 |
|
1005 - 1926 |
921 |
HGTFTCASEYT-PFKLTCATTRQ |
0.01038 |
|
1015 - 1029 |
14 |
TGNYQCGHYKH-KETLYCIDGAL |
0.02216 |
|
1015 - 1071 |
56 |
TGNYQCGHYKH-LDGVVCTEIDP |
0.0105 |
|
1015 - 1114 |
99 |
TGNYQCGHYKH-NFKFVCDNIKF |
0.01041 |
|
1015 - 1191 |
176 |
TGNYQCGHYKH-KPNTWCIRCLW |
0.0104 |
|
1015 - 1194 |
179 |
TGNYQCGHYKH-TWCIRCLWSTK |
0.10065 |
|
1015 - 1223 |
208 |
TGNYQCGHYKH-MDNLACEDLKP |
0.40779 |
|
1015 - 1246 |
231 |
TGNYQCGHYKH-KDVLECNVKTT |
0.99756 |
|
1015 - 1342 |
327 |
TGNYQCGHYKH-NIVTRCLNRVC |
0.01277 |
|
1015 - 1347 |
332 |
TGNYQCGHYKH-CLNRVCTNYMP |
0.01043 |
|
1015 - 1362 |
347 |
TGNYQCGHYKH-LLLQLCTFTRS |
0.01088 |
|
1015 - 1392 |
377 |
TGNYQCGHYKH-SVGKFCLEASF |
0.01066 |
|
1015 - 1421 |
406 |
TGNYQCGHYKH-LLLSVCLGSLI |
0.01083 |
|
1015 - 1445 |
430 |
TGNYQCGHYKH-GMPSYCTGYRE |
0.01056 |
|
1015 - 1464 |
449 |
TGNYQCGHYKH-TIATYCTGSIP |
0.99962 |
|
1015 - 1470 |
455 |
TGNYQCGHYKH-TGSIPCSVCLS |
0.01044 |
|
1015 - 1473 |
458 |
TGNYQCGHYKH-IPCSVCLSGLD |
0.96091 |
|
1015 - 1586 |
571 |
TGNYQCGHYKH-HVVDGCNSSTC |
0.99624 |
|
1015 - 1591 |
576 |
TGNYQCGHYKH-CNSSTCMMCYK |
0.01037 |
|
1015 - 1594 |
579 |
TGNYQCGHYKH-STCMMCYKRNR |
0.82162 |
|
1015 - 1605 |
590 |
TGNYQCGHYKH-ATRVECTTIVN |
0.01042 |
|
1015 - 1627 |
612 |
TGNYQCGHYKH-GGKGFCKLHNW |
0.41412 |
|
1015 - 1634 |
619 |
TGNYQCGHYKH-LHNWNCVNCDT |
0.99581 |
|
1015 - 1637 |
622 |
TGNYQCGHYKH-WNCVNCDTFCA |
0.99898 |
|
1015 - 1641 |
626 |
TGNYQCGHYKH-NCDTFCAGSTF |
0.01125 |
|
1015 - 1731 |
716 |
TGNYQCGHYKH-DGKSKCEESSA |
0.01037 |
|
1015 - 1748 |
733 |
TGNYQCGHYKH-YSQLMCQPILL |
0.01062 |
|
1015 - 1836 |
821 |
TGNYQCGHYKH-KDVVECLKLSH |
0.0106 |
|
1015 - 1852 |
837 |
TGNYQCGHYKH-VTGDSCNNYML |
0.01125 |
|
1015 - 1873 |
858 |
TGNYQCGHYKH-RDLGACIDCSA |
0.01093 |
|
1015 - 1876 |
861 |
TGNYQCGHYKH-GACIDCSARHI |
0.99476 |
|
1015 - 1926 |
911 |
TGNYQCGHYKH-PFKLTCATTRQ |
0.01053 |
|
1029 - 1071 |
42 |
KETLYCIDGAL-LDGVVCTEIDP |
0.0118 |
|
1029 - 1114 |
85 |
KETLYCIDGAL-NFKFVCDNIKF |
0.01383 |
|
1029 - 1191 |
162 |
KETLYCIDGAL-KPNTWCIRCLW |
0.0104 |
|
1029 - 1194 |
165 |
KETLYCIDGAL-TWCIRCLWSTK |
0.01047 |
|
1029 - 1223 |
194 |
KETLYCIDGAL-MDNLACEDLKP |
0.01764 |
|
1029 - 1246 |
217 |
KETLYCIDGAL-KDVLECNVKTT |
0.01047 |
|
1029 - 1342 |
313 |
KETLYCIDGAL-NIVTRCLNRVC |
0.01038 |
|
1029 - 1347 |
318 |
KETLYCIDGAL-CLNRVCTNYMP |
0.01314 |
|
1029 - 1362 |
333 |
KETLYCIDGAL-LLLQLCTFTRS |
0.01278 |
|
1029 - 1392 |
363 |
KETLYCIDGAL-SVGKFCLEASF |
0.01042 |
|
1029 - 1421 |
392 |
KETLYCIDGAL-LLLSVCLGSLI |
0.01214 |
|
1029 - 1445 |
416 |
KETLYCIDGAL-GMPSYCTGYRE |
0.01049 |
|
1029 - 1464 |
435 |
KETLYCIDGAL-TIATYCTGSIP |
0.01113 |
|
1029 - 1470 |
441 |
KETLYCIDGAL-TGSIPCSVCLS |
0.01279 |
|
1029 - 1473 |
444 |
KETLYCIDGAL-IPCSVCLSGLD |
0.01079 |
|
1029 - 1586 |
557 |
KETLYCIDGAL-HVVDGCNSSTC |
0.01038 |
|
1029 - 1591 |
562 |
KETLYCIDGAL-CNSSTCMMCYK |
0.01038 |
|
1029 - 1594 |
565 |
KETLYCIDGAL-STCMMCYKRNR |
0.01041 |
|
1029 - 1605 |
576 |
KETLYCIDGAL-ATRVECTTIVN |
0.01677 |
|
1029 - 1627 |
598 |
KETLYCIDGAL-GGKGFCKLHNW |
0.12146 |
|
1029 - 1634 |
605 |
KETLYCIDGAL-LHNWNCVNCDT |
0.01044 |
|
1029 - 1637 |
608 |
KETLYCIDGAL-WNCVNCDTFCA |
0.01052 |
|
1029 - 1641 |
612 |
KETLYCIDGAL-NCDTFCAGSTF |
0.99904 |
|
1029 - 1731 |
702 |
KETLYCIDGAL-DGKSKCEESSA |
0.01213 |
|
1029 - 1748 |
719 |
KETLYCIDGAL-YSQLMCQPILL |
0.02934 |
|
1029 - 1836 |
807 |
KETLYCIDGAL-KDVVECLKLSH |
0.0104 |
|
1029 - 1852 |
823 |
KETLYCIDGAL-VTGDSCNNYML |
0.01231 |
|
1029 - 1873 |
844 |
KETLYCIDGAL-RDLGACIDCSA |
0.01104 |
|
1029 - 1876 |
847 |
KETLYCIDGAL-GACIDCSARHI |
0.0106 |
|
1029 - 1926 |
897 |
KETLYCIDGAL-PFKLTCATTRQ |
0.01038 |
|
1071 - 1114 |
43 |
LDGVVCTEIDP-NFKFVCDNIKF |
0.01042 |
|
1071 - 1191 |
120 |
LDGVVCTEIDP-KPNTWCIRCLW |
0.01051 |
|
1071 - 1194 |
123 |
LDGVVCTEIDP-TWCIRCLWSTK |
0.01039 |
|
1071 - 1223 |
152 |
LDGVVCTEIDP-MDNLACEDLKP |
0.01059 |
|
1071 - 1246 |
175 |
LDGVVCTEIDP-KDVLECNVKTT |
0.01055 |
|
1071 - 1342 |
271 |
LDGVVCTEIDP-NIVTRCLNRVC |
0.01038 |
|
1071 - 1347 |
276 |
LDGVVCTEIDP-CLNRVCTNYMP |
0.01037 |
|
1071 - 1362 |
291 |
LDGVVCTEIDP-LLLQLCTFTRS |
0.01039 |
|
1071 - 1392 |
321 |
LDGVVCTEIDP-SVGKFCLEASF |
0.01037 |
|
1071 - 1421 |
350 |
LDGVVCTEIDP-LLLSVCLGSLI |
0.01037 |
|
1071 - 1445 |
374 |
LDGVVCTEIDP-GMPSYCTGYRE |
0.01039 |
|
1071 - 1464 |
393 |
LDGVVCTEIDP-TIATYCTGSIP |
0.0127 |
|
1071 - 1470 |
399 |
LDGVVCTEIDP-TGSIPCSVCLS |
0.01038 |
|
1071 - 1473 |
402 |
LDGVVCTEIDP-IPCSVCLSGLD |
0.01039 |
|
1071 - 1586 |
515 |
LDGVVCTEIDP-HVVDGCNSSTC |
0.01045 |
|
1071 - 1591 |
520 |
LDGVVCTEIDP-CNSSTCMMCYK |
0.01037 |
|
1071 - 1594 |
523 |
LDGVVCTEIDP-STCMMCYKRNR |
0.01128 |
|
1071 - 1605 |
534 |
LDGVVCTEIDP-ATRVECTTIVN |
0.01041 |
|
1071 - 1627 |
556 |
LDGVVCTEIDP-GGKGFCKLHNW |
0.01536 |
|
1071 - 1634 |
563 |
LDGVVCTEIDP-LHNWNCVNCDT |
0.01043 |
|
1071 - 1637 |
566 |
LDGVVCTEIDP-WNCVNCDTFCA |
0.01104 |
|
1071 - 1641 |
570 |
LDGVVCTEIDP-NCDTFCAGSTF |
0.01044 |
|
1071 - 1731 |
660 |
LDGVVCTEIDP-DGKSKCEESSA |
0.01038 |
|
1071 - 1748 |
677 |
LDGVVCTEIDP-YSQLMCQPILL |
0.01041 |
|
1071 - 1836 |
765 |
LDGVVCTEIDP-KDVVECLKLSH |
0.01037 |
|
1071 - 1852 |
781 |
LDGVVCTEIDP-VTGDSCNNYML |
0.01039 |
|
1071 - 1873 |
802 |
LDGVVCTEIDP-RDLGACIDCSA |
0.01041 |
|
1071 - 1876 |
805 |
LDGVVCTEIDP-GACIDCSARHI |
0.01092 |
|
1071 - 1926 |
855 |
LDGVVCTEIDP-PFKLTCATTRQ |
0.01037 |
|
1114 - 1191 |
77 |
NFKFVCDNIKF-KPNTWCIRCLW |
0.99729 |
|
1114 - 1194 |
80 |
NFKFVCDNIKF-TWCIRCLWSTK |
0.95189 |
|
1114 - 1223 |
109 |
NFKFVCDNIKF-MDNLACEDLKP |
0.07304 |
|
1114 - 1246 |
132 |
NFKFVCDNIKF-KDVLECNVKTT |
0.99679 |
|
1114 - 1342 |
228 |
NFKFVCDNIKF-NIVTRCLNRVC |
0.01336 |
|
1114 - 1347 |
233 |
NFKFVCDNIKF-CLNRVCTNYMP |
0.02396 |
|
1114 - 1362 |
248 |
NFKFVCDNIKF-LLLQLCTFTRS |
0.99958 |
|
1114 - 1392 |
278 |
NFKFVCDNIKF-SVGKFCLEASF |
0.01533 |
|
1114 - 1421 |
307 |
NFKFVCDNIKF-LLLSVCLGSLI |
0.99836 |
|
1114 - 1445 |
331 |
NFKFVCDNIKF-GMPSYCTGYRE |
0.99832 |
|
1114 - 1464 |
350 |
NFKFVCDNIKF-TIATYCTGSIP |
0.99695 |
|
1114 - 1470 |
356 |
NFKFVCDNIKF-TGSIPCSVCLS |
0.99671 |
|
1114 - 1473 |
359 |
NFKFVCDNIKF-IPCSVCLSGLD |
0.01039 |
|
1114 - 1586 |
472 |
NFKFVCDNIKF-HVVDGCNSSTC |
0.05243 |
|
1114 - 1591 |
477 |
NFKFVCDNIKF-CNSSTCMMCYK |
0.98088 |
|
1114 - 1594 |
480 |
NFKFVCDNIKF-STCMMCYKRNR |
0.99729 |
|
1114 - 1605 |
491 |
NFKFVCDNIKF-ATRVECTTIVN |
0.99695 |
|
1114 - 1627 |
513 |
NFKFVCDNIKF-GGKGFCKLHNW |
0.99735 |
|
1114 - 1634 |
520 |
NFKFVCDNIKF-LHNWNCVNCDT |
0.99707 |
|
1114 - 1637 |
523 |
NFKFVCDNIKF-WNCVNCDTFCA |
0.99716 |
|
1114 - 1641 |
527 |
NFKFVCDNIKF-NCDTFCAGSTF |
0.99867 |
|
1114 - 1731 |
617 |
NFKFVCDNIKF-DGKSKCEESSA |
0.99738 |
|
1114 - 1748 |
634 |
NFKFVCDNIKF-YSQLMCQPILL |
0.99836 |
|
1114 - 1836 |
722 |
NFKFVCDNIKF-KDVVECLKLSH |
0.01385 |
|
1114 - 1852 |
738 |
NFKFVCDNIKF-VTGDSCNNYML |
0.99731 |
|
1114 - 1873 |
759 |
NFKFVCDNIKF-RDLGACIDCSA |
0.05932 |
|
1114 - 1876 |
762 |
NFKFVCDNIKF-GACIDCSARHI |
0.99737 |
|
1114 - 1926 |
812 |
NFKFVCDNIKF-PFKLTCATTRQ |
0.98052 |
|
1191 - 1194 |
3 |
KPNTWCIRCLW-TWCIRCLWSTK |
0.01126 |
|
1191 - 1223 |
32 |
KPNTWCIRCLW-MDNLACEDLKP |
0.01088 |
|
1191 - 1246 |
55 |
KPNTWCIRCLW-KDVLECNVKTT |
0.01833 |
|
1191 - 1342 |
151 |
KPNTWCIRCLW-NIVTRCLNRVC |
0.0104 |
|
1191 - 1347 |
156 |
KPNTWCIRCLW-CLNRVCTNYMP |
0.01039 |
|
1191 - 1362 |
171 |
KPNTWCIRCLW-LLLQLCTFTRS |
0.01038 |
|
1191 - 1392 |
201 |
KPNTWCIRCLW-SVGKFCLEASF |
0.01038 |
|
1191 - 1421 |
230 |
KPNTWCIRCLW-LLLSVCLGSLI |
0.01037 |
|
1191 - 1445 |
254 |
KPNTWCIRCLW-GMPSYCTGYRE |
0.01057 |
|
1191 - 1464 |
273 |
KPNTWCIRCLW-TIATYCTGSIP |
0.01123 |
|
1191 - 1470 |
279 |
KPNTWCIRCLW-TGSIPCSVCLS |
0.78931 |
|
1191 - 1473 |
282 |
KPNTWCIRCLW-IPCSVCLSGLD |
0.01361 |
|
1191 - 1586 |
395 |
KPNTWCIRCLW-HVVDGCNSSTC |
0.04219 |
|
1191 - 1591 |
400 |
KPNTWCIRCLW-CNSSTCMMCYK |
0.01132 |
|
1191 - 1594 |
403 |
KPNTWCIRCLW-STCMMCYKRNR |
0.01068 |
|
1191 - 1605 |
414 |
KPNTWCIRCLW-ATRVECTTIVN |
0.01059 |
|
1191 - 1627 |
436 |
KPNTWCIRCLW-GGKGFCKLHNW |
0.01042 |
|
1191 - 1634 |
443 |
KPNTWCIRCLW-LHNWNCVNCDT |
0.01812 |
|
1191 - 1637 |
446 |
KPNTWCIRCLW-WNCVNCDTFCA |
0.14953 |
|
1191 - 1641 |
450 |
KPNTWCIRCLW-NCDTFCAGSTF |
0.99048 |
|
1191 - 1731 |
540 |
KPNTWCIRCLW-DGKSKCEESSA |
0.01183 |
|
1191 - 1748 |
557 |
KPNTWCIRCLW-YSQLMCQPILL |
0.9729 |
|
1191 - 1836 |
645 |
KPNTWCIRCLW-KDVVECLKLSH |
0.01189 |
|
1191 - 1852 |
661 |
KPNTWCIRCLW-VTGDSCNNYML |
0.99719 |
|
1191 - 1873 |
682 |
KPNTWCIRCLW-RDLGACIDCSA |
0.01038 |
|
1191 - 1876 |
685 |
KPNTWCIRCLW-GACIDCSARHI |
0.01119 |
|
1191 - 1926 |
735 |
KPNTWCIRCLW-PFKLTCATTRQ |
0.01128 |
|
1194 - 1223 |
29 |
TWCIRCLWSTK-MDNLACEDLKP |
0.01038 |
|
1194 - 1246 |
52 |
TWCIRCLWSTK-KDVLECNVKTT |
0.01038 |
|
1194 - 1342 |
148 |
TWCIRCLWSTK-NIVTRCLNRVC |
0.01037 |
|
1194 - 1347 |
153 |
TWCIRCLWSTK-CLNRVCTNYMP |
0.01037 |
|
1194 - 1362 |
168 |
TWCIRCLWSTK-LLLQLCTFTRS |
0.0104 |
|
1194 - 1392 |
198 |
TWCIRCLWSTK-SVGKFCLEASF |
0.01037 |
|
1194 - 1421 |
227 |
TWCIRCLWSTK-LLLSVCLGSLI |
0.01037 |
|
1194 - 1445 |
251 |
TWCIRCLWSTK-GMPSYCTGYRE |
0.01045 |
|
1194 - 1464 |
270 |
TWCIRCLWSTK-TIATYCTGSIP |
0.01038 |
|
1194 - 1470 |
276 |
TWCIRCLWSTK-TGSIPCSVCLS |
0.0135 |
|
1194 - 1473 |
279 |
TWCIRCLWSTK-IPCSVCLSGLD |
0.01038 |
|
1194 - 1586 |
392 |
TWCIRCLWSTK-HVVDGCNSSTC |
0.01038 |
|
1194 - 1591 |
397 |
TWCIRCLWSTK-CNSSTCMMCYK |
0.01039 |
|
1194 - 1594 |
400 |
TWCIRCLWSTK-STCMMCYKRNR |
0.01235 |
|
1194 - 1605 |
411 |
TWCIRCLWSTK-ATRVECTTIVN |
0.01037 |
|
1194 - 1627 |
433 |
TWCIRCLWSTK-GGKGFCKLHNW |
0.13936 |
|
1194 - 1634 |
440 |
TWCIRCLWSTK-LHNWNCVNCDT |
0.01039 |
|
1194 - 1637 |
443 |
TWCIRCLWSTK-WNCVNCDTFCA |
0.01041 |
|
1194 - 1641 |
447 |
TWCIRCLWSTK-NCDTFCAGSTF |
0.91367 |
|
1194 - 1731 |
537 |
TWCIRCLWSTK-DGKSKCEESSA |
0.01062 |
|
1194 - 1748 |
554 |
TWCIRCLWSTK-YSQLMCQPILL |
0.15434 |
|
1194 - 1836 |
642 |
TWCIRCLWSTK-KDVVECLKLSH |
0.01046 |
|
1194 - 1852 |
658 |
TWCIRCLWSTK-VTGDSCNNYML |
0.01163 |
|
1194 - 1873 |
679 |
TWCIRCLWSTK-RDLGACIDCSA |
0.01037 |
|
1194 - 1876 |
682 |
TWCIRCLWSTK-GACIDCSARHI |
0.01044 |
|
1194 - 1926 |
732 |
TWCIRCLWSTK-PFKLTCATTRQ |
0.01038 |
|
1223 - 1246 |
23 |
MDNLACEDLKP-KDVLECNVKTT |
0.01062 |
|
1223 - 1342 |
119 |
MDNLACEDLKP-NIVTRCLNRVC |
0.01037 |
|
1223 - 1347 |
124 |
MDNLACEDLKP-CLNRVCTNYMP |
0.01038 |
|
1223 - 1362 |
139 |
MDNLACEDLKP-LLLQLCTFTRS |
0.01037 |
|
1223 - 1392 |
169 |
MDNLACEDLKP-SVGKFCLEASF |
0.01037 |
|
1223 - 1421 |
198 |
MDNLACEDLKP-LLLSVCLGSLI |
0.01037 |
|
1223 - 1445 |
222 |
MDNLACEDLKP-GMPSYCTGYRE |
0.86776 |
|
1223 - 1464 |
241 |
MDNLACEDLKP-TIATYCTGSIP |
0.01074 |
|
1223 - 1470 |
247 |
MDNLACEDLKP-TGSIPCSVCLS |
0.01123 |
|
1223 - 1473 |
250 |
MDNLACEDLKP-IPCSVCLSGLD |
0.01038 |
|
1223 - 1586 |
363 |
MDNLACEDLKP-HVVDGCNSSTC |
0.01038 |
|
1223 - 1591 |
368 |
MDNLACEDLKP-CNSSTCMMCYK |
0.01148 |
|
1223 - 1594 |
371 |
MDNLACEDLKP-STCMMCYKRNR |
0.99558 |
|
1223 - 1605 |
382 |
MDNLACEDLKP-ATRVECTTIVN |
0.01111 |
|
1223 - 1627 |
404 |
MDNLACEDLKP-GGKGFCKLHNW |
0.99721 |
|
1223 - 1634 |
411 |
MDNLACEDLKP-LHNWNCVNCDT |
0.01045 |
|
1223 - 1637 |
414 |
MDNLACEDLKP-WNCVNCDTFCA |
0.01078 |
|
1223 - 1641 |
418 |
MDNLACEDLKP-NCDTFCAGSTF |
0.01057 |
|
1223 - 1731 |
508 |
MDNLACEDLKP-DGKSKCEESSA |
0.28876 |
|
1223 - 1748 |
525 |
MDNLACEDLKP-YSQLMCQPILL |
0.0105 |
|
1223 - 1836 |
613 |
MDNLACEDLKP-KDVVECLKLSH |
0.01037 |
|
1223 - 1852 |
629 |
MDNLACEDLKP-VTGDSCNNYML |
0.0171 |
|
1223 - 1873 |
650 |
MDNLACEDLKP-RDLGACIDCSA |
0.01037 |
|
1223 - 1876 |
653 |
MDNLACEDLKP-GACIDCSARHI |
0.01072 |
|
1223 - 1926 |
703 |
MDNLACEDLKP-PFKLTCATTRQ |
0.01056 |
|
1246 - 1342 |
96 |
KDVLECNVKTT-NIVTRCLNRVC |
0.01039 |
|
1246 - 1347 |
101 |
KDVLECNVKTT-CLNRVCTNYMP |
0.0104 |
|
1246 - 1362 |
116 |
KDVLECNVKTT-LLLQLCTFTRS |
0.01038 |
|
1246 - 1392 |
146 |
KDVLECNVKTT-SVGKFCLEASF |
0.01037 |
|
1246 - 1421 |
175 |
KDVLECNVKTT-LLLSVCLGSLI |
0.01037 |
|
1246 - 1445 |
199 |
KDVLECNVKTT-GMPSYCTGYRE |
0.99572 |
|
1246 - 1464 |
218 |
KDVLECNVKTT-TIATYCTGSIP |
0.05648 |
|
1246 - 1470 |
224 |
KDVLECNVKTT-TGSIPCSVCLS |
0.52491 |
|
1246 - 1473 |
227 |
KDVLECNVKTT-IPCSVCLSGLD |
0.01276 |
|
1246 - 1586 |
340 |
KDVLECNVKTT-HVVDGCNSSTC |
0.01078 |
|
1246 - 1591 |
345 |
KDVLECNVKTT-CNSSTCMMCYK |
0.01152 |
|
1246 - 1594 |
348 |
KDVLECNVKTT-STCMMCYKRNR |
0.99721 |
|
1246 - 1605 |
359 |
KDVLECNVKTT-ATRVECTTIVN |
0.01768 |
|
1246 - 1627 |
381 |
KDVLECNVKTT-GGKGFCKLHNW |
0.99729 |
|
1246 - 1634 |
388 |
KDVLECNVKTT-LHNWNCVNCDT |
0.02408 |
|
1246 - 1637 |
391 |
KDVLECNVKTT-WNCVNCDTFCA |
0.41161 |
|
1246 - 1641 |
395 |
KDVLECNVKTT-NCDTFCAGSTF |
0.04958 |
|
1246 - 1731 |
485 |
KDVLECNVKTT-DGKSKCEESSA |
0.95465 |
|
1246 - 1748 |
502 |
KDVLECNVKTT-YSQLMCQPILL |
0.65331 |
|
1246 - 1836 |
590 |
KDVLECNVKTT-KDVVECLKLSH |
0.01118 |
|
1246 - 1852 |
606 |
KDVLECNVKTT-VTGDSCNNYML |
0.9997 |
|
1246 - 1873 |
627 |
KDVLECNVKTT-RDLGACIDCSA |
0.01037 |
|
1246 - 1876 |
630 |
KDVLECNVKTT-GACIDCSARHI |
0.24485 |
|
1246 - 1926 |
680 |
KDVLECNVKTT-PFKLTCATTRQ |
0.01184 |
|
1342 - 1347 |
5 |
NIVTRCLNRVC-CLNRVCTNYMP |
0.28257 |
|
1342 - 1362 |
20 |
NIVTRCLNRVC-LLLQLCTFTRS |
0.052 |
|
1342 - 1392 |
50 |
NIVTRCLNRVC-SVGKFCLEASF |
0.01538 |
|
1342 - 1421 |
79 |
NIVTRCLNRVC-LLLSVCLGSLI |
0.01149 |
|
1342 - 1445 |
103 |
NIVTRCLNRVC-GMPSYCTGYRE |
0.01042 |
|
1342 - 1464 |
122 |
NIVTRCLNRVC-TIATYCTGSIP |
0.01037 |
|
1342 - 1470 |
128 |
NIVTRCLNRVC-TGSIPCSVCLS |
0.57621 |
|
1342 - 1473 |
131 |
NIVTRCLNRVC-IPCSVCLSGLD |
0.2482 |
|
1342 - 1586 |
244 |
NIVTRCLNRVC-HVVDGCNSSTC |
0.01042 |
|
1342 - 1591 |
249 |
NIVTRCLNRVC-CNSSTCMMCYK |
0.0104 |
|
1342 - 1594 |
252 |
NIVTRCLNRVC-STCMMCYKRNR |
0.01452 |
|
1342 - 1605 |
263 |
NIVTRCLNRVC-ATRVECTTIVN |
0.07578 |
|
1342 - 1627 |
285 |
NIVTRCLNRVC-GGKGFCKLHNW |
0.01293 |
|
1342 - 1634 |
292 |
NIVTRCLNRVC-LHNWNCVNCDT |
0.01062 |
|
1342 - 1637 |
295 |
NIVTRCLNRVC-WNCVNCDTFCA |
0.01091 |
|
1342 - 1641 |
299 |
NIVTRCLNRVC-NCDTFCAGSTF |
0.96899 |
|
1342 - 1731 |
389 |
NIVTRCLNRVC-DGKSKCEESSA |
0.01053 |
|
1342 - 1748 |
406 |
NIVTRCLNRVC-YSQLMCQPILL |
0.12837 |
|
1342 - 1836 |
494 |
NIVTRCLNRVC-KDVVECLKLSH |
0.01139 |
|
1342 - 1852 |
510 |
NIVTRCLNRVC-VTGDSCNNYML |
0.29152 |
|
1342 - 1873 |
531 |
NIVTRCLNRVC-RDLGACIDCSA |
0.01224 |
|
1342 - 1876 |
534 |
NIVTRCLNRVC-GACIDCSARHI |
0.01039 |
|
1342 - 1926 |
584 |
NIVTRCLNRVC-PFKLTCATTRQ |
0.0104 |
|
1347 - 1362 |
15 |
CLNRVCTNYMP-LLLQLCTFTRS |
0.01042 |
|
1347 - 1392 |
45 |
CLNRVCTNYMP-SVGKFCLEASF |
0.01038 |
|
1347 - 1421 |
74 |
CLNRVCTNYMP-LLLSVCLGSLI |
0.01037 |
|
1347 - 1445 |
98 |
CLNRVCTNYMP-GMPSYCTGYRE |
0.01048 |
|
1347 - 1464 |
117 |
CLNRVCTNYMP-TIATYCTGSIP |
0.01039 |
|
1347 - 1470 |
123 |
CLNRVCTNYMP-TGSIPCSVCLS |
0.04564 |
|
1347 - 1473 |
126 |
CLNRVCTNYMP-IPCSVCLSGLD |
0.01111 |
|
1347 - 1586 |
239 |
CLNRVCTNYMP-HVVDGCNSSTC |
0.01039 |
|
1347 - 1591 |
244 |
CLNRVCTNYMP-CNSSTCMMCYK |
0.0104 |
|
1347 - 1594 |
247 |
CLNRVCTNYMP-STCMMCYKRNR |
0.01037 |
|
1347 - 1605 |
258 |
CLNRVCTNYMP-ATRVECTTIVN |
0.01096 |
|
1347 - 1627 |
280 |
CLNRVCTNYMP-GGKGFCKLHNW |
0.0104 |
|
1347 - 1634 |
287 |
CLNRVCTNYMP-LHNWNCVNCDT |
0.01041 |
|
1347 - 1637 |
290 |
CLNRVCTNYMP-WNCVNCDTFCA |
0.01097 |
|
1347 - 1641 |
294 |
CLNRVCTNYMP-NCDTFCAGSTF |
0.24593 |
|
1347 - 1731 |
384 |
CLNRVCTNYMP-DGKSKCEESSA |
0.01046 |
|
1347 - 1748 |
401 |
CLNRVCTNYMP-YSQLMCQPILL |
0.0134 |
|
1347 - 1836 |
489 |
CLNRVCTNYMP-KDVVECLKLSH |
0.01046 |
|
1347 - 1852 |
505 |
CLNRVCTNYMP-VTGDSCNNYML |
0.04128 |
|
1347 - 1873 |
526 |
CLNRVCTNYMP-RDLGACIDCSA |
0.01038 |
|
1347 - 1876 |
529 |
CLNRVCTNYMP-GACIDCSARHI |
0.01038 |
|
1347 - 1926 |
579 |
CLNRVCTNYMP-PFKLTCATTRQ |
0.01037 |
|
1362 - 1392 |
30 |
LLLQLCTFTRS-SVGKFCLEASF |
0.01072 |
|
1362 - 1421 |
59 |
LLLQLCTFTRS-LLLSVCLGSLI |
0.01041 |
|
1362 - 1445 |
83 |
LLLQLCTFTRS-GMPSYCTGYRE |
0.0104 |
|
1362 - 1464 |
102 |
LLLQLCTFTRS-TIATYCTGSIP |
0.0106 |
|
1362 - 1470 |
108 |
LLLQLCTFTRS-TGSIPCSVCLS |
0.04317 |
|
1362 - 1473 |
111 |
LLLQLCTFTRS-IPCSVCLSGLD |
0.03466 |
|
1362 - 1586 |
224 |
LLLQLCTFTRS-HVVDGCNSSTC |
0.01047 |
|
1362 - 1591 |
229 |
LLLQLCTFTRS-CNSSTCMMCYK |
0.01045 |
|
1362 - 1594 |
232 |
LLLQLCTFTRS-STCMMCYKRNR |
0.01043 |
|
1362 - 1605 |
243 |
LLLQLCTFTRS-ATRVECTTIVN |
0.03247 |
|
1362 - 1627 |
265 |
LLLQLCTFTRS-GGKGFCKLHNW |
0.01085 |
|
1362 - 1634 |
272 |
LLLQLCTFTRS-LHNWNCVNCDT |
0.01045 |
|
1362 - 1637 |
275 |
LLLQLCTFTRS-WNCVNCDTFCA |
0.01098 |
|
1362 - 1641 |
279 |
LLLQLCTFTRS-NCDTFCAGSTF |
0.04387 |
|
1362 - 1731 |
369 |
LLLQLCTFTRS-DGKSKCEESSA |
0.01058 |
|
1362 - 1748 |
386 |
LLLQLCTFTRS-YSQLMCQPILL |
0.34243 |
|
1362 - 1836 |
474 |
LLLQLCTFTRS-KDVVECLKLSH |
0.01063 |
|
1362 - 1852 |
490 |
LLLQLCTFTRS-VTGDSCNNYML |
0.35499 |
|
1362 - 1873 |
511 |
LLLQLCTFTRS-RDLGACIDCSA |
0.01098 |
|
1362 - 1876 |
514 |
LLLQLCTFTRS-GACIDCSARHI |
0.01051 |
|
1362 - 1926 |
564 |
LLLQLCTFTRS-PFKLTCATTRQ |
0.0104 |
|
1392 - 1421 |
29 |
SVGKFCLEASF-LLLSVCLGSLI |
0.01037 |
|
1392 - 1445 |
53 |
SVGKFCLEASF-GMPSYCTGYRE |
0.01093 |
|
1392 - 1464 |
72 |
SVGKFCLEASF-TIATYCTGSIP |
0.01039 |
|
1392 - 1470 |
78 |
SVGKFCLEASF-TGSIPCSVCLS |
0.90011 |
|
1392 - 1473 |
81 |
SVGKFCLEASF-IPCSVCLSGLD |
0.01305 |
|
1392 - 1586 |
194 |
SVGKFCLEASF-HVVDGCNSSTC |
0.01089 |
|
1392 - 1591 |
199 |
SVGKFCLEASF-CNSSTCMMCYK |
0.01061 |
|
1392 - 1594 |
202 |
SVGKFCLEASF-STCMMCYKRNR |
0.01041 |
|
1392 - 1605 |
213 |
SVGKFCLEASF-ATRVECTTIVN |
0.01097 |
|
1392 - 1627 |
235 |
SVGKFCLEASF-GGKGFCKLHNW |
0.01044 |
|
1392 - 1634 |
242 |
SVGKFCLEASF-LHNWNCVNCDT |
0.01144 |
|
1392 - 1637 |
245 |
SVGKFCLEASF-WNCVNCDTFCA |
0.01497 |
|
1392 - 1641 |
249 |
SVGKFCLEASF-NCDTFCAGSTF |
0.99916 |
|
1392 - 1731 |
339 |
SVGKFCLEASF-DGKSKCEESSA |
0.01219 |
|
1392 - 1748 |
356 |
SVGKFCLEASF-YSQLMCQPILL |
0.94345 |
|
1392 - 1836 |
444 |
SVGKFCLEASF-KDVVECLKLSH |
0.01117 |
|
1392 - 1852 |
460 |
SVGKFCLEASF-VTGDSCNNYML |
0.99494 |
|
1392 - 1873 |
481 |
SVGKFCLEASF-RDLGACIDCSA |
0.01044 |
|
1392 - 1876 |
484 |
SVGKFCLEASF-GACIDCSARHI |
0.01061 |
|
1392 - 1926 |
534 |
SVGKFCLEASF-PFKLTCATTRQ |
0.01045 |
|
1421 - 1445 |
24 |
LLLSVCLGSLI-GMPSYCTGYRE |
0.01037 |
|
1421 - 1464 |
43 |
LLLSVCLGSLI-TIATYCTGSIP |
0.01038 |
|
1421 - 1470 |
49 |
LLLSVCLGSLI-TGSIPCSVCLS |
0.01275 |
|
1421 - 1473 |
52 |
LLLSVCLGSLI-IPCSVCLSGLD |
0.98934 |
|
1421 - 1586 |
165 |
LLLSVCLGSLI-HVVDGCNSSTC |
0.01038 |
|
1421 - 1591 |
170 |
LLLSVCLGSLI-CNSSTCMMCYK |
0.01037 |
|
1421 - 1594 |
173 |
LLLSVCLGSLI-STCMMCYKRNR |
0.01188 |
|
1421 - 1605 |
184 |
LLLSVCLGSLI-ATRVECTTIVN |
0.98595 |
|
1421 - 1627 |
206 |
LLLSVCLGSLI-GGKGFCKLHNW |
0.01038 |
|
1421 - 1634 |
213 |
LLLSVCLGSLI-LHNWNCVNCDT |
0.01427 |
|
1421 - 1637 |
216 |
LLLSVCLGSLI-WNCVNCDTFCA |
0.01042 |
|
1421 - 1641 |
220 |
LLLSVCLGSLI-NCDTFCAGSTF |
0.03987 |
|
1421 - 1731 |
310 |
LLLSVCLGSLI-DGKSKCEESSA |
0.01039 |
|
1421 - 1748 |
327 |
LLLSVCLGSLI-YSQLMCQPILL |
0.07691 |
|
1421 - 1836 |
415 |
LLLSVCLGSLI-KDVVECLKLSH |
0.04576 |
|
1421 - 1852 |
431 |
LLLSVCLGSLI-VTGDSCNNYML |
0.01536 |
|
1421 - 1873 |
452 |
LLLSVCLGSLI-RDLGACIDCSA |
0.23973 |
|
1421 - 1876 |
455 |
LLLSVCLGSLI-GACIDCSARHI |
0.01037 |
|
1421 - 1926 |
505 |
LLLSVCLGSLI-PFKLTCATTRQ |
0.01087 |
|
1445 - 1464 |
19 |
GMPSYCTGYRE-TIATYCTGSIP |
0.84139 |
|
1445 - 1470 |
25 |
GMPSYCTGYRE-TGSIPCSVCLS |
0.01111 |
|
1445 - 1473 |
28 |
GMPSYCTGYRE-IPCSVCLSGLD |
0.03314 |
|
1445 - 1586 |
141 |
GMPSYCTGYRE-HVVDGCNSSTC |
0.01171 |
|
1445 - 1591 |
146 |
GMPSYCTGYRE-CNSSTCMMCYK |
0.01037 |
|
1445 - 1594 |
149 |
GMPSYCTGYRE-STCMMCYKRNR |
0.01181 |
|
1445 - 1605 |
160 |
GMPSYCTGYRE-ATRVECTTIVN |
0.01069 |
|
1445 - 1627 |
182 |
GMPSYCTGYRE-GGKGFCKLHNW |
0.01042 |
|
1445 - 1634 |
189 |
GMPSYCTGYRE-LHNWNCVNCDT |
0.01102 |
|
1445 - 1637 |
192 |
GMPSYCTGYRE-WNCVNCDTFCA |
0.02755 |
|
1445 - 1641 |
196 |
GMPSYCTGYRE-NCDTFCAGSTF |
0.01086 |
|
1445 - 1731 |
286 |
GMPSYCTGYRE-DGKSKCEESSA |
0.01037 |
|
1445 - 1748 |
303 |
GMPSYCTGYRE-YSQLMCQPILL |
0.01084 |
|
1445 - 1836 |
391 |
GMPSYCTGYRE-KDVVECLKLSH |
0.01057 |
|
1445 - 1852 |
407 |
GMPSYCTGYRE-VTGDSCNNYML |
0.01087 |
|
1445 - 1873 |
428 |
GMPSYCTGYRE-RDLGACIDCSA |
0.03762 |
|
1445 - 1876 |
431 |
GMPSYCTGYRE-GACIDCSARHI |
0.95164 |
|
1445 - 1926 |
481 |
GMPSYCTGYRE-PFKLTCATTRQ |
0.01041 |
|
1464 - 1470 |
6 |
TIATYCTGSIP-TGSIPCSVCLS |
0.01102 |
|
1464 - 1473 |
9 |
TIATYCTGSIP-IPCSVCLSGLD |
0.01055 |
|
1464 - 1586 |
122 |
TIATYCTGSIP-HVVDGCNSSTC |
0.01039 |
|
1464 - 1591 |
127 |
TIATYCTGSIP-CNSSTCMMCYK |
0.01038 |
|
1464 - 1594 |
130 |
TIATYCTGSIP-STCMMCYKRNR |
0.99437 |
|
1464 - 1605 |
141 |
TIATYCTGSIP-ATRVECTTIVN |
0.01056 |
|
1464 - 1627 |
163 |
TIATYCTGSIP-GGKGFCKLHNW |
0.99719 |
|
1464 - 1634 |
170 |
TIATYCTGSIP-LHNWNCVNCDT |
0.01115 |
|
1464 - 1637 |
173 |
TIATYCTGSIP-WNCVNCDTFCA |
0.01172 |
|
1464 - 1641 |
177 |
TIATYCTGSIP-NCDTFCAGSTF |
0.01283 |
|
1464 - 1731 |
267 |
TIATYCTGSIP-DGKSKCEESSA |
0.01083 |
|
1464 - 1748 |
284 |
TIATYCTGSIP-YSQLMCQPILL |
0.01072 |
|
1464 - 1836 |
372 |
TIATYCTGSIP-KDVVECLKLSH |
0.01039 |
|
1464 - 1852 |
388 |
TIATYCTGSIP-VTGDSCNNYML |
0.27525 |
|
1464 - 1873 |
409 |
TIATYCTGSIP-RDLGACIDCSA |
0.01056 |
|
1464 - 1876 |
412 |
TIATYCTGSIP-GACIDCSARHI |
0.01155 |
|
1464 - 1926 |
462 |
TIATYCTGSIP-PFKLTCATTRQ |
0.01044 |
|
1470 - 1473 |
3 |
TGSIPCSVCLS-IPCSVCLSGLD |
0.01072 |
|
1470 - 1586 |
116 |
TGSIPCSVCLS-HVVDGCNSSTC |
0.01038 |
|
1470 - 1591 |
121 |
TGSIPCSVCLS-CNSSTCMMCYK |
0.01037 |
|
1470 - 1594 |
124 |
TGSIPCSVCLS-STCMMCYKRNR |
0.01568 |
|
1470 - 1605 |
135 |
TGSIPCSVCLS-ATRVECTTIVN |
0.01048 |
|
1470 - 1627 |
157 |
TGSIPCSVCLS-GGKGFCKLHNW |
0.15447 |
|
1470 - 1634 |
164 |
TGSIPCSVCLS-LHNWNCVNCDT |
0.01049 |
|
1470 - 1637 |
167 |
TGSIPCSVCLS-WNCVNCDTFCA |
0.01042 |
|
1470 - 1641 |
171 |
TGSIPCSVCLS-NCDTFCAGSTF |
0.01173 |
|
1470 - 1731 |
261 |
TGSIPCSVCLS-DGKSKCEESSA |
0.0104 |
|
1470 - 1748 |
278 |
TGSIPCSVCLS-YSQLMCQPILL |
0.01044 |
|
1470 - 1836 |
366 |
TGSIPCSVCLS-KDVVECLKLSH |
0.01037 |
|
1470 - 1852 |
382 |
TGSIPCSVCLS-VTGDSCNNYML |
0.01045 |
|
1470 - 1873 |
403 |
TGSIPCSVCLS-RDLGACIDCSA |
0.0105 |
|
1470 - 1876 |
406 |
TGSIPCSVCLS-GACIDCSARHI |
0.01164 |
|
1470 - 1926 |
456 |
TGSIPCSVCLS-PFKLTCATTRQ |
0.01037 |
|
1473 - 1586 |
113 |
IPCSVCLSGLD-HVVDGCNSSTC |
0.01038 |
|
1473 - 1591 |
118 |
IPCSVCLSGLD-CNSSTCMMCYK |
0.01924 |
|
1473 - 1594 |
121 |
IPCSVCLSGLD-STCMMCYKRNR |
0.99714 |
|
1473 - 1605 |
132 |
IPCSVCLSGLD-ATRVECTTIVN |
0.03066 |
|
1473 - 1627 |
154 |
IPCSVCLSGLD-GGKGFCKLHNW |
0.99726 |
|
1473 - 1634 |
161 |
IPCSVCLSGLD-LHNWNCVNCDT |
0.01071 |
|
1473 - 1637 |
164 |
IPCSVCLSGLD-WNCVNCDTFCA |
0.0178 |
|
1473 - 1641 |
168 |
IPCSVCLSGLD-NCDTFCAGSTF |
0.01107 |
|
1473 - 1731 |
258 |
IPCSVCLSGLD-DGKSKCEESSA |
0.23244 |
|
1473 - 1748 |
275 |
IPCSVCLSGLD-YSQLMCQPILL |
0.01416 |
|
1473 - 1836 |
363 |
IPCSVCLSGLD-KDVVECLKLSH |
0.0104 |
|
1473 - 1852 |
379 |
IPCSVCLSGLD-VTGDSCNNYML |
0.88891 |
|
1473 - 1873 |
400 |
IPCSVCLSGLD-RDLGACIDCSA |
0.01121 |
|
1473 - 1876 |
403 |
IPCSVCLSGLD-GACIDCSARHI |
0.02966 |
|
1473 - 1926 |
453 |
IPCSVCLSGLD-PFKLTCATTRQ |
0.01074 |
|
1586 - 1591 |
5 |
HVVDGCNSSTC-CNSSTCMMCYK |
0.01039 |
|
1586 - 1594 |
8 |
HVVDGCNSSTC-STCMMCYKRNR |
0.93587 |
|
1586 - 1605 |
19 |
HVVDGCNSSTC-ATRVECTTIVN |
0.02313 |
|
1586 - 1627 |
41 |
HVVDGCNSSTC-GGKGFCKLHNW |
0.9967 |
|
1586 - 1634 |
48 |
HVVDGCNSSTC-LHNWNCVNCDT |
0.0117 |
|
1586 - 1637 |
51 |
HVVDGCNSSTC-WNCVNCDTFCA |
0.01141 |
|
1586 - 1641 |
55 |
HVVDGCNSSTC-NCDTFCAGSTF |
0.99908 |
|
1586 - 1731 |
145 |
HVVDGCNSSTC-DGKSKCEESSA |
0.01084 |
|
1586 - 1748 |
162 |
HVVDGCNSSTC-YSQLMCQPILL |
0.01058 |
|
1586 - 1836 |
250 |
HVVDGCNSSTC-KDVVECLKLSH |
0.01039 |
|
1586 - 1852 |
266 |
HVVDGCNSSTC-VTGDSCNNYML |
0.09478 |
|
1586 - 1873 |
287 |
HVVDGCNSSTC-RDLGACIDCSA |
0.02677 |
|
1586 - 1876 |
290 |
HVVDGCNSSTC-GACIDCSARHI |
0.01068 |
|
1586 - 1926 |
340 |
HVVDGCNSSTC-PFKLTCATTRQ |
0.0104 |
|
1591 - 1594 |
3 |
CNSSTCMMCYK-STCMMCYKRNR |
0.72096 |
|
1591 - 1605 |
14 |
CNSSTCMMCYK-ATRVECTTIVN |
0.01078 |
|
1591 - 1627 |
36 |
CNSSTCMMCYK-GGKGFCKLHNW |
0.99698 |
|
1591 - 1634 |
43 |
CNSSTCMMCYK-LHNWNCVNCDT |
0.01103 |
|
1591 - 1637 |
46 |
CNSSTCMMCYK-WNCVNCDTFCA |
0.01042 |
|
1591 - 1641 |
50 |
CNSSTCMMCYK-NCDTFCAGSTF |
0.01062 |
|
1591 - 1731 |
140 |
CNSSTCMMCYK-DGKSKCEESSA |
0.01384 |
|
1591 - 1748 |
157 |
CNSSTCMMCYK-YSQLMCQPILL |
0.01041 |
|
1591 - 1836 |
245 |
CNSSTCMMCYK-KDVVECLKLSH |
0.01037 |
|
1591 - 1852 |
261 |
CNSSTCMMCYK-VTGDSCNNYML |
0.0179 |
|
1591 - 1873 |
282 |
CNSSTCMMCYK-RDLGACIDCSA |
0.01037 |
|
1591 - 1876 |
285 |
CNSSTCMMCYK-GACIDCSARHI |
0.76446 |
|
1591 - 1926 |
335 |
CNSSTCMMCYK-PFKLTCATTRQ |
0.0104 |
|
1594 - 1605 |
11 |
STCMMCYKRNR-ATRVECTTIVN |
0.0104 |
|
1594 - 1627 |
33 |
STCMMCYKRNR-GGKGFCKLHNW |
0.01231 |
|
1594 - 1634 |
40 |
STCMMCYKRNR-LHNWNCVNCDT |
0.01037 |
|
1594 - 1637 |
43 |
STCMMCYKRNR-WNCVNCDTFCA |
0.01037 |
|
1594 - 1641 |
47 |
STCMMCYKRNR-NCDTFCAGSTF |
0.01249 |
|
1594 - 1731 |
137 |
STCMMCYKRNR-DGKSKCEESSA |
0.01037 |
|
1594 - 1748 |
154 |
STCMMCYKRNR-YSQLMCQPILL |
0.01049 |
|
1594 - 1836 |
242 |
STCMMCYKRNR-KDVVECLKLSH |
0.01037 |
|
1594 - 1852 |
258 |
STCMMCYKRNR-VTGDSCNNYML |
0.01039 |
|
1594 - 1873 |
279 |
STCMMCYKRNR-RDLGACIDCSA |
0.01038 |
|
1594 - 1876 |
282 |
STCMMCYKRNR-GACIDCSARHI |
0.01037 |
|
1594 - 1926 |
332 |
STCMMCYKRNR-PFKLTCATTRQ |
0.01037 |
|
1605 - 1627 |
22 |
ATRVECTTIVN-GGKGFCKLHNW |
0.99728 |
|
1605 - 1634 |
29 |
ATRVECTTIVN-LHNWNCVNCDT |
0.06908 |
|
1605 - 1637 |
32 |
ATRVECTTIVN-WNCVNCDTFCA |
0.02753 |
|
1605 - 1641 |
36 |
ATRVECTTIVN-NCDTFCAGSTF |
0.53652 |
|
1605 - 1731 |
126 |
ATRVECTTIVN-DGKSKCEESSA |
0.35269 |
|
1605 - 1748 |
143 |
ATRVECTTIVN-YSQLMCQPILL |
0.50279 |
|
1605 - 1836 |
231 |
ATRVECTTIVN-KDVVECLKLSH |
0.01158 |
|
1605 - 1852 |
247 |
ATRVECTTIVN-VTGDSCNNYML |
0.99573 |
|
1605 - 1873 |
268 |
ATRVECTTIVN-RDLGACIDCSA |
0.01037 |
|
1605 - 1876 |
271 |
ATRVECTTIVN-GACIDCSARHI |
0.66758 |
|
1605 - 1926 |
321 |
ATRVECTTIVN-PFKLTCATTRQ |
0.0117 |
|
1627 - 1634 |
7 |
GGKGFCKLHNW-LHNWNCVNCDT |
0.01045 |
|
1627 - 1637 |
10 |
GGKGFCKLHNW-WNCVNCDTFCA |
0.01039 |
|
1627 - 1641 |
14 |
GGKGFCKLHNW-NCDTFCAGSTF |
0.01188 |
|
1627 - 1731 |
104 |
GGKGFCKLHNW-DGKSKCEESSA |
0.01037 |
|
1627 - 1748 |
121 |
GGKGFCKLHNW-YSQLMCQPILL |
0.01038 |
|
1627 - 1836 |
209 |
GGKGFCKLHNW-KDVVECLKLSH |
0.01037 |
|
1627 - 1852 |
225 |
GGKGFCKLHNW-VTGDSCNNYML |
0.01043 |
|
1627 - 1873 |
246 |
GGKGFCKLHNW-RDLGACIDCSA |
0.01056 |
|
1627 - 1876 |
249 |
GGKGFCKLHNW-GACIDCSARHI |
0.0106 |
|
1627 - 1926 |
299 |
GGKGFCKLHNW-PFKLTCATTRQ |
0.01037 |
|
1634 - 1637 |
3 |
LHNWNCVNCDT-WNCVNCDTFCA |
0.01056 |
|
1634 - 1641 |
7 |
LHNWNCVNCDT-NCDTFCAGSTF |
0.01129 |
|
1634 - 1731 |
97 |
LHNWNCVNCDT-DGKSKCEESSA |
0.01037 |
|
1634 - 1748 |
114 |
LHNWNCVNCDT-YSQLMCQPILL |
0.01384 |
|
1634 - 1836 |
202 |
LHNWNCVNCDT-KDVVECLKLSH |
0.01038 |
|
1634 - 1852 |
218 |
LHNWNCVNCDT-VTGDSCNNYML |
0.01064 |
|
1634 - 1873 |
239 |
LHNWNCVNCDT-RDLGACIDCSA |
0.01159 |
|
1634 - 1876 |
242 |
LHNWNCVNCDT-GACIDCSARHI |
0.01105 |
|
1634 - 1926 |
292 |
LHNWNCVNCDT-PFKLTCATTRQ |
0.01038 |
|
1637 - 1641 |
4 |
WNCVNCDTFCA-NCDTFCAGSTF |
0.01061 |
|
1637 - 1731 |
94 |
WNCVNCDTFCA-DGKSKCEESSA |
0.01037 |
|
1637 - 1748 |
111 |
WNCVNCDTFCA-YSQLMCQPILL |
0.01043 |
|
1637 - 1836 |
199 |
WNCVNCDTFCA-KDVVECLKLSH |
0.01039 |
|
1637 - 1852 |
215 |
WNCVNCDTFCA-VTGDSCNNYML |
0.01052 |
|
1637 - 1873 |
236 |
WNCVNCDTFCA-RDLGACIDCSA |
0.01052 |
|
1637 - 1876 |
239 |
WNCVNCDTFCA-GACIDCSARHI |
0.02117 |
|
1637 - 1926 |
289 |
WNCVNCDTFCA-PFKLTCATTRQ |
0.01041 |
|
1641 - 1731 |
90 |
NCDTFCAGSTF-DGKSKCEESSA |
0.01194 |
|
1641 - 1748 |
107 |
NCDTFCAGSTF-YSQLMCQPILL |
0.0738 |
|
1641 - 1836 |
195 |
NCDTFCAGSTF-KDVVECLKLSH |
0.01079 |
|
1641 - 1852 |
211 |
NCDTFCAGSTF-VTGDSCNNYML |
0.01337 |
|
1641 - 1873 |
232 |
NCDTFCAGSTF-RDLGACIDCSA |
0.46484 |
|
1641 - 1876 |
235 |
NCDTFCAGSTF-GACIDCSARHI |
0.01046 |
|
1641 - 1926 |
285 |
NCDTFCAGSTF-PFKLTCATTRQ |
0.01076 |
|
1731 - 1748 |
17 |
DGKSKCEESSA-YSQLMCQPILL |
0.01242 |
|
1731 - 1836 |
105 |
DGKSKCEESSA-KDVVECLKLSH |
0.01037 |
|
1731 - 1852 |
121 |
DGKSKCEESSA-VTGDSCNNYML |
0.97574 |
|
1731 - 1873 |
142 |
DGKSKCEESSA-RDLGACIDCSA |
0.01038 |
|
1731 - 1876 |
145 |
DGKSKCEESSA-GACIDCSARHI |
0.02076 |
|
1731 - 1926 |
195 |
DGKSKCEESSA-PFKLTCATTRQ |
0.01203 |
|
1748 - 1836 |
88 |
YSQLMCQPILL-KDVVECLKLSH |
0.01048 |
|
1748 - 1852 |
104 |
YSQLMCQPILL-VTGDSCNNYML |
0.01739 |
|
1748 - 1873 |
125 |
YSQLMCQPILL-RDLGACIDCSA |
0.01038 |
|
1748 - 1876 |
128 |
YSQLMCQPILL-GACIDCSARHI |
0.01037 |
|
1748 - 1926 |
178 |
YSQLMCQPILL-PFKLTCATTRQ |
0.01039 |
|
1836 - 1852 |
16 |
KDVVECLKLSH-VTGDSCNNYML |
0.97837 |
|
1836 - 1873 |
37 |
KDVVECLKLSH-RDLGACIDCSA |
0.01039 |
|
1836 - 1876 |
40 |
KDVVECLKLSH-GACIDCSARHI |
0.01045 |
|
1836 - 1926 |
90 |
KDVVECLKLSH-PFKLTCATTRQ |
0.01041 |
|
1852 - 1873 |
21 |
VTGDSCNNYML-RDLGACIDCSA |
0.01037 |
|
1852 - 1876 |
24 |
VTGDSCNNYML-GACIDCSARHI |
0.01046 |
|
1852 - 1926 |
74 |
VTGDSCNNYML-PFKLTCATTRQ |
0.01039 |
|
1873 - 1876 |
3 |
RDLGACIDCSA-GACIDCSARHI |
0.01038 |
|
1873 - 1926 |
53 |
RDLGACIDCSA-PFKLTCATTRQ |
0.01038 |
|
1876 - 1926 |
50 |
GACIDCSARHI-PFKLTCATTRQ |
0.01037 |