Comparison between Conventional qPCR and Microfluidic Chip-Based PCR System for COVID-19 Nucleic Acid Detection
Article Information
Francesca Dragoni1,2, Maria Garofalo1,2, Rosa Trotti3, Yiteng Liu4, Cristina Cereda1,*, Stella Gagliardi1
1Genomic and post-Genomic Unit, IRCCS Mondino Foundation, Pavia, Italy
2Department of Biology and Biotechnology "L. Spallanzani", University of Pavia, Pavia, Italy
3Department of Neurodiagnostics and Services, Laboratory of Clinicals and Chemicals Analysis (SMeL), IRCCS Mondino Foundation
4The Earth, Ocean and Atmospheric Sciences Thrust, The Hong Kong University of Science and Technol- ogy, Clear Water Bay, Kowloon, Hong Kong, China
*Corresponding Author: Cristina Cereda, Genomic and post-Genomic Unit, IRCCS Mondino Foundation, Pavia, Italy
Received: 01 December 2021; Accepted: 07 December 2021; Published: 29 December 2021
Citation:
Francesca Dragoni, Maria Garofalo, Rosa Trotti, Yiteng Liu, Cristina Cereda, Stella Gagliardi. Comparison between Conventional qPCR and Microfluidic Chip-based PCR System for COVID-19 Nucleic Acid Detection. Journal of Psychiatry and Psychiatric Disorders 5 (2021): 218-231.
Share at FacebookAbstract
In the last year, COVID-19, rapidly evolved in a global pandemic. Clinical diagnosis is fundamental for the restriction of pandemic spread and different approaches have been proposed. Since large-scale laboratory screening takes time to ensure sensitivity of diagnostic tests, aim of this study was to suggest a fast and sensitive viral nucleic acids’ detection system from nasopharyngeal swabs collected in Universal Transport Media (UTM). We compare the common qRT-PCR technique with the innovative SWM-01 method to detect viral RNA from 20 samples collected in UTM. We also provide a new qRT-PCR protocol based on the use of lower amount of reagents in the reaction mix useful to avoid waste of material in pandemic time. We demonstrate that results obtained with two different detection systems and different amount of reagents are comparable. Moreover, we demonstrate that strong positive samples are correctly detected with the new system and with higher sensitivity than the classic qRT-PCR which detects samples with a delay. Weakly positive samples, instead, are incorrectly assigned with the new analyzer possibly due to a detection limit of the instrument itself. We propose the SHINEWAY SWM-01 Nucleic Acids Analyzer (SWM-01) as a promising diagnostic system suitable for routine diagnosis process.
Keywords
COVID-19; SARS-CoV-2; Pandemic; Swabs; qRT-PCR; SWM-01 Analyzer
COVID-19 articles; SARS-CoV-2 articles; Pandemic articles; Swabs articles; qRT-PCR articles; SWM-01 Analyzer articles
COVID-19 articles COVID-19 Research articles COVID-19 review articles COVID-19 PubMed articles COVID-19 PubMed Central articles COVID-19 2023 articles COVID-19 2024 articles COVID-19 Scopus articles COVID-19 impact factor journals COVID-19 Scopus journals COVID-19 PubMed journals COVID-19 medical journals COVID-19 free journals COVID-19 best journals COVID-19 top journals COVID-19 free medical journals COVID-19 famous journals COVID-19 Google Scholar indexed journals SARS-CoV-2 articles SARS-CoV-2 Research articles SARS-CoV-2 review articles SARS-CoV-2 PubMed articles SARS-CoV-2 PubMed Central articles SARS-CoV-2 2023 articles SARS-CoV-2 2024 articles SARS-CoV-2 Scopus articles SARS-CoV-2 impact factor journals SARS-CoV-2 Scopus journals SARS-CoV-2 PubMed journals SARS-CoV-2 medical journals SARS-CoV-2 free journals SARS-CoV-2 best journals SARS-CoV-2 top journals SARS-CoV-2 free medical journals SARS-CoV-2 famous journals SARS-CoV-2 Google Scholar indexed journals Pandemic articles Pandemic Research articles Pandemic review articles Pandemic PubMed articles Pandemic PubMed Central articles Pandemic 2023 articles Pandemic 2024 articles Pandemic Scopus articles Pandemic impact factor journals Pandemic Scopus journals Pandemic PubMed journals Pandemic medical journals Pandemic free journals Pandemic best journals Pandemic top journals Pandemic free medical journals Pandemic famous journals Pandemic Google Scholar indexed journals Swabs articles Swabs Research articles Swabs review articles Swabs PubMed articles Swabs PubMed Central articles Swabs 2023 articles Swabs 2024 articles Swabs Scopus articles Swabs impact factor journals Swabs Scopus journals Swabs PubMed journals Swabs medical journals Swabs free journals Swabs best journals Swabs top journals Swabs free medical journals Swabs famous journals Swabs Google Scholar indexed journals qRT-PCR articles qRT-PCR Research articles qRT-PCR review articles qRT-PCR PubMed articles qRT-PCR PubMed Central articles qRT-PCR 2023 articles qRT-PCR 2024 articles qRT-PCR Scopus articles qRT-PCR impact factor journals qRT-PCR Scopus journals qRT-PCR PubMed journals qRT-PCR medical journals qRT-PCR free journals qRT-PCR best journals qRT-PCR top journals qRT-PCR free medical journals qRT-PCR famous journals qRT-PCR Google Scholar indexed journals SWM-01 Analyzer articles SWM-01 Analyzer Research articles SWM-01 Analyzer review articles SWM-01 Analyzer PubMed articles SWM-01 Analyzer PubMed Central articles SWM-01 Analyzer 2023 articles SWM-01 Analyzer 2024 articles SWM-01 Analyzer Scopus articles SWM-01 Analyzer impact factor journals SWM-01 Analyzer Scopus journals SWM-01 Analyzer PubMed journals SWM-01 Analyzer medical journals SWM-01 Analyzer free journals SWM-01 Analyzer best journals SWM-01 Analyzer top journals SWM-01 Analyzer free medical journals SWM-01 Analyzer famous journals SWM-01 Analyzer Google Scholar indexed journals Universal Transport Media articles Universal Transport Media Research articles Universal Transport Media review articles Universal Transport Media PubMed articles Universal Transport Media PubMed Central articles Universal Transport Media 2023 articles Universal Transport Media 2024 articles Universal Transport Media Scopus articles Universal Transport Media impact factor journals Universal Transport Media Scopus journals Universal Transport Media PubMed journals Universal Transport Media medical journals Universal Transport Media free journals Universal Transport Media best journals Universal Transport Media top journals Universal Transport Media free medical journals Universal Transport Media famous journals Universal Transport Media Google Scholar indexed journals RNA polymerase articles RNA polymerase Research articles RNA polymerase review articles RNA polymerase PubMed articles RNA polymerase PubMed Central articles RNA polymerase 2023 articles RNA polymerase 2024 articles RNA polymerase Scopus articles RNA polymerase impact factor journals RNA polymerase Scopus journals RNA polymerase PubMed journals RNA polymerase medical journals RNA polymerase free journals RNA polymerase best journals RNA polymerase top journals RNA polymerase free medical journals RNA polymerase famous journals RNA polymerase Google Scholar indexed journals Point-of-care Testing articles Point-of-care Testing Research articles Point-of-care Testing review articles Point-of-care Testing PubMed articles Point-of-care Testing PubMed Central articles Point-of-care Testing 2023 articles Point-of-care Testing 2024 articles Point-of-care Testing Scopus articles Point-of-care Testing impact factor journals Point-of-care Testing Scopus journals Point-of-care Testing PubMed journals Point-of-care Testing medical journals Point-of-care Testing free journals Point-of-care Testing best journals Point-of-care Testing top journals Point-of-care Testing free medical journals Point-of-care Testing famous journals Point-of-care Testing Google Scholar indexed journals CT scan articles CT scan Research articles CT scan review articles CT scan PubMed articles CT scan PubMed Central articles CT scan 2023 articles CT scan 2024 articles CT scan Scopus articles CT scan impact factor journals CT scan Scopus journals CT scan PubMed journals CT scan medical journals CT scan free journals CT scan best journals CT scan top journals CT scan free medical journals CT scan famous journals CT scan Google Scholar indexed journals
Article Details
1. Introduction
Coronavirus disease-19 (COVID-19) is a pneumonia caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). During the last year, after the outbreak of SARS-CoV-2 in China, COVID-19 rapidly evolved in a global pandemic [1, 2]. The virus is part of the Coronaviridae family, which include enveloped, positive-sense single-stranded RNA viruses. Its genome shows similarities with the genome of SARS-CoV and MERS-CoV and encodes for sixteen nonstructural proteins including RNA-dependent RNA polymerase (RdRp), four structural proteins: spike (S), envelope (E), membrane (M), nucleocapsid (N), and nine accessory proteins involved in replication processes [2-6]. With a very short incubation period of 5-6 days, SARS-CoV-2 cause a flu-like disease with common symptoms including fever, cough, fatigue, dyspnea, shortness of breath, acute respiratory failure and, occasionally, gastrointestinal dysfunctions [5, 7-10]. Clinical diagnosis of COVID-19 is fundamental for the restriction of pandemic spread and many different approaches have been approved as nucleic acids isolation and detection, CT scan, Biosensor, immune identification technology, Point-of-care Testing (POCT) of IgM/IgG, enzyme-linked immunosorbent assay (ELISA), indirect fluorescent antibody techniques, electrochemical sensors and blood culture too [5, 8, 11, 12]. Usually, specimens for diagnostic test are obtained through oropharyngeal swabs or alternatively through nasopharyngeal swabs [13]. RNA genome extraction allows detection and eventually quantification of viral charge. The most common techniques for nucleic acids detection are the quantitative Real Time RT-PCR (qRT-PCR) and high-throughput sequencing. Several qRT-PCR test kits have been commercialized, based on the use of specific primers and probes for SARS-CoV-2 gene regions that encode for the nucleocapsid (N), envelope (E), spike (S), and RNA-dependent RNA polymerase proteins (RdRP) [10]. Although qRT-PCR provides adequate sensitivity for the detection of early infection, one of the most important clinical aspects for the pandemic spread is the risk of false-negative or false-positive results [14]. In case of low concentration of the virus, the sensitivity of analysis could not be enough to detect viral nucleic acids, especially in asymptomatic or weakly positive patients [5]. Moreover, the timing of diagnosis is crucial to prevent incorrect results [15]. In addition, as large-scale laboratory screening takes time to ensure reliability and sensitivity of diagnostic tests, and the increased request for laboratory analysis has led companies to lack of diagnostic products; several new approaches and instruments have been proposed during this time.
Here we report the new SHINEWAY SWM-01 Analyzer (SWM-01) (Simitecno, IT) as a possible solution for easier and faster analysis. The innovative SWM-01 PCR Nucleic Acids Analyzer performs qualitative detection of nucleic acids, both DNA and RNA, by single fluorescence PCR. The product can be used in various scenarios including CDC, medical treatment emergency, specialist visit, primary care and blood screening. The analyzer is made up of the instrument and the power supply, while the tool is comprised of the control system, the power supply system, the photoelectric system, the temperature control system, the case component, the reaction chip, and software modules. The device uses optical detection and has three fluorescence channels. The microfluidic chip is made up of three parallel channels and sits on top of a temperature-controlled heater. The microfluidic chip's pooling technology allows it to analyze up to 9 samples at the same time, in 45 minutes, excluding the execution of the swab and the RNA extraction. Recently, microfluidic technology has been applied in several research field including high throughput screening. Microfluidic devices are useful not only for saving space but also because miniaturization offers different superiorities [16]. Differently from a classical qRT-PCR System, the SWM-01 Analyzer with the pooling technology can test a minimum of 3 patients in 50 minutes and a maximum of 18 patients in 90 minutes. This item offers an advanced optical path design to eliminate external light interference and further increase detection reliability; moreover an accurate and efficient data processing system with an artificial intelligence image segmentation algorithm is available. The timing required for the process is less than traditional technologies which usually take from 2 to 3 hours. A micro-heater is used as a rapid heating component, and the microfluidic chip is used as a support for PCR amplification. The PCR amplification system use a fluorescent probe specific for the detection of SARS-CoV-2 viral RNA. During the PCR detection, the fluorescence of positive samples accumulates and the signal is collected as images from a CMOS camera. Finally, the control system completes the data collection and provide, as a final result, the positivity or negativity of samples. The system also provides a quantification of the viral RNA detected expressed as cycle threshold (Ct).
Aim of this study was to optimize a new quantitative Real-Time PCR protocol based on the use of the SWM-01 Analyzer with lower amount of reagents, to compare results between the most common Real-Time PCR and the innovative SWM-01 method and to suggest a faster and easier approach to detect viral RNA from nasopharyngeal swabs.
2. Materials and Methods
2.1 RNA extraction
RNA from nasopharyngeal swabs collected in UTM have been extracted with the automated benchtop nucleic acids extraction system NUCLISENS® EASYMAG® (Biomérieux, IT) according to manufacturer’s specifications. 20 samples were extracted as described above and analyzed, including 8 negative samples, 11 positive samples, one synthetic COVID-19 positive control and one synthetic negative control. This study was developed on existing samples collected during standard diagnostic test for SARS-CoV-2 RNA detection. Synthetic positive and negative controls were provided with the Novel Coronavirus 2019-nCoV nucleic acid Detection kit (BioFlux) used in the qRT-PCR reaction.
2.2 Quantitative Real Time PCR
The Real-Time PCR was performed using the SARS-CoV-2, Novel Coronavirus 2019-nCoV nucleic acid Detection kit (BioFlux). In this assay we used a standard reaction mix volume of 25 µL (12.5 µL RT-PCR Buffer Master Mix, 1.3 µL RT-PCR Enzyme Mix, 6.2 µL Probe Mix with 5 μL of extracted RNA) and a reduced reaction mix volume of 15 µL (6.1 µL RT-PCR Buffer Master Mix, 0.8 µL RT-PCR Enzyme Mix, 3.1 µL Probe Mix with 5 µL of extracted RNA). The Real-Time PCR protocol included 10 minutes at 50°C, 1 minute at 95°C and 30 seconds at 60°C repeated for 45 cycles. The one-step fluorescence RT-PCR reaction system allowed us to perform retrotrascription of viral RNA directly during the qRT-PCR amplification. Two detection channel, TEXAS RED 610 and FAM, were used to amplify ORF1ab and N gene respectively. Ct values has been recorded for each sample. Ct<40 correspond to positive samples and Ct>35 correspond to weak positive ones. To perform this analysis, the CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) has been used.
2.3 SWM-01 quantitative Real Time PCR
As previously described for the classical qRT-PCR, the reaction was performed using the SARS-CoV-2, Novel Coronavirus 2019-nCoV nucleic acid Detection kit (BioFlux), comparing results from two reaction mix volume of 25 µL (12.5 µL RT-PCR Buffer Master Mix, 1.3 µL RT-PCR Enzyme Mix, 6.2 µL Probe Mix with 5 μL of extracted RNA) and 15 µL (6.1 µL RT-PCR Buffer Master Mix, 0.8 µL RT-PCR Enzyme Mix, 3.1 µL Probe Mix with 5 µL of extracted RNA), the volume of the microchip is 12 μL in both cases. The Real-Time PCR protocol included 10 minutes at 50°C, 1 minute at 95°C and 10 seconds at 95°C, 30 seconds at 60°C (ON) repeated for 45 cycles. Ct values has been recorded for each sample. Ct<40 correspond to positive samples and Ct>35 correspond to weak positive ones. To perform this analysis the on-site PCR System SHINEWAY SWM-01 PCR Nucleic Acids Analyzer (Simitecno, IT) has been used.
3. Results and Discussion
3.1 Optimization of SWM-01 qReal-Time PCR protocol
In the first part of this work, we tried to optimize the qRT-PCR protocol in order to reduce the amount of reagents used in the reaction. We tested the same COVID-19 positive and negative controls, both with a standard reaction mix volume of 25 µL and reduced reaction mix volume of 15 µL, in order to assess if this parameter could interfere with the reaction efficiency. Additionally, we performed qRT-PCR with two different instruments, comparing results obtained from a classical CFX96™ Real-Time PCR Detection System and from the new SWM-01 Analyzer. We demonstrate that the Ct values recorded for the samples processed with the standard reaction mix volume of 25 µL are comparable with the ones recorded for the same samples with the decreased reaction mix volume of 15 µL; same results were obtained with both instruments (Figure 1).
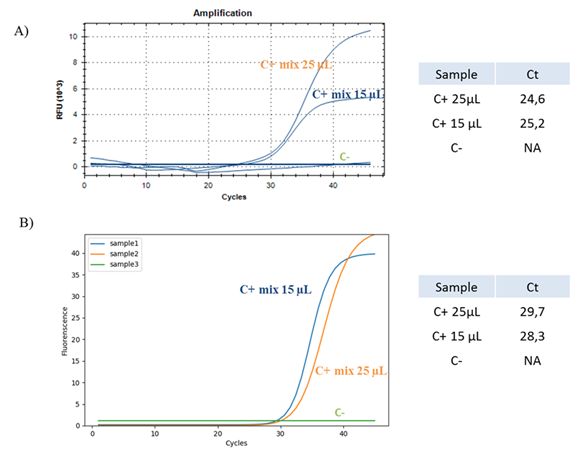
Figure 1:Optimization of Real Time-PCR protocol. (A) Real Time-PCR of one synthetic COVID-19 positive control (blue and orange) and one synthetic COVID-19 negative control (green) with two different reaction mix volumes of 15 µL (blue) and 25 µL (orange) performed with a classical detection system. RFU is the Relative Fluorescence Unit. (B) Real Time-PCR of one COVID-19 positive control (blue and orange) and one COVID-19 negative control (green) with two different reaction mix volumes of 15 µL (blue) and 25 µL (orange) performed with the SWM-01 Analyzer. C+ is the synthetic positive control, C- is the synthetic negative control. C+ mix 15 uL is the synthetic positive control processed with a mix of 15 uL. C+ mix 25 uL is the synthetic positive control processed with a mix of 25 uL. Ct is the Cycle Threshold value.
Our data demonstrate that the outcomes obtained from two different detection systems and with different amount of reagents are comparable, suggesting the opportunity to avoid useless waste of materials.
3.2 Comparison between common qRT-PCR and innovative SWM-01 method
Since the results obtained in the first part of our work demonstrate that the SWM-01 analyzer provides same outcomes as a classical RT-PCR Detection System, it was investigated if it was possible to use this new instrument with an advantage for the sensitivity and for the timing of large scale COVID-19 screening.
3.2.1 Synthetic COVID-19 positive sample: Firstly, in order to verify the sensitivity of the instrument, we tested a synthetic COVID-19 positive sample at two different dilutions (1:2 and 1:5), comparing results obtained from classical CFX96™ Real Time PCR Detection system and from the SWM-01 Analyzer (Figure 2) with a reaction mix volume of 15 ul.
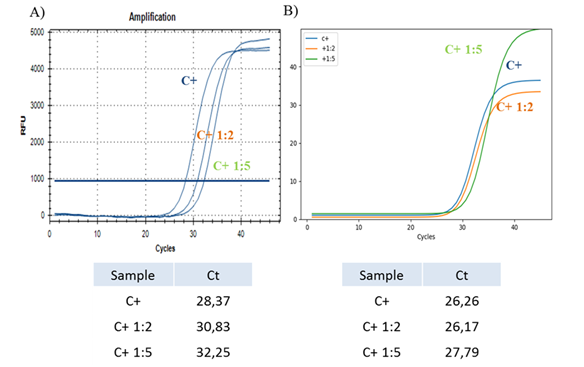
Figure 2: Real-Time PCR with dilute samples. (A) Real-Time PCR of one synthetic COVID-19 positive control at initial concentration (blue) and 1:2 (orange), 1:5 (green) dilutions performed with a classical detection system. The table shows the relative Ct values for each sample. (B) Real-Time PCR of one synthetic COVID-19 positive control at initial concentration (blue) and 1:2 (orange), 1:5 (green) dilutions performed with the SWM-01 Analyzer and relative Ct values for each sample.
Our data show that the new SWM-01 Analyzer could perform Real-Time PCR with higher sensitivity for the viral nucleic acids detection in dilute samples than the traditional qRT-PCR techniques, since samples analyzed with the classical CFX96™ Real-Time PCR Detection system exhibit Ct values with a ‘delay’ of two and four cycles compared to the Ct values recorded for the samples analyzed with the new method. Moreover, the 1:2 diluted sample analyzed with the SWM-01 instrument, show the same Ct values of the sample at the initial concentration and after the 1:5 dilution, same sample is detected with a ‘delay’ of 1.5 cycles.
3.2.2 UTM samples
|
Sample |
Bio-rad (Bioer kit) Ct |
SWM-01 (Bioer kit) Ct |
|
28320 |
25 |
28 |
|
28169 |
24 |
27 |
|
28314 |
28 |
NA |
|
28196 |
30 |
34 |
|
13194 |
31 |
NA |
|
13195 |
32 |
38 |
|
56182 |
26 |
28 |
|
13196 |
25 |
29 |
|
13153 |
29 |
32 |
|
13155 |
26 |
28 |
|
13178 |
36 |
NA |
|
C+ |
28 |
26 |
|
Negative samples=8 |
NA |
NA |
Table 1: Summary of samples derived from nasopharyngeal swabs and collected in UTM, analyzed with the SWM-01 Analyzer and with CFX96™ Real-Time PCR Detection system.
In order to confirm these data, we performed a RT-PCR with 20 samples (12 positive and 8 negative samples) obtained from nasopharyngeal swabs and collected in UTM (Table 1) with a reaction mix volume of 15 uL.
3.2.2.1 Positive samples in UTM: In this first part we report data from three positive samples as example. The assay was performed with both instruments to compare their viral RNA detection sensitivity (Figure 3).
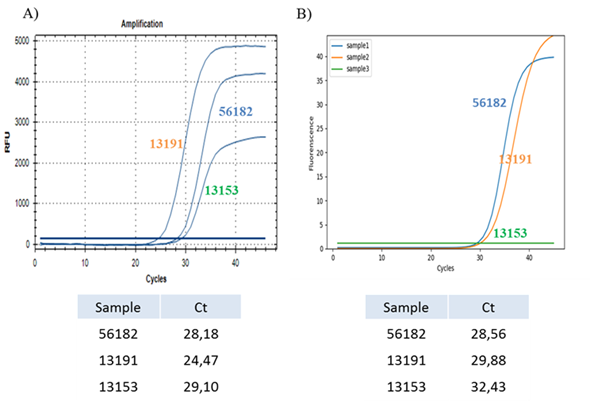
Figure 3: Comparison between common Real-Time PCR amplification and innovative SWM-01 method on samples collected in UTM. (A) Real-Time PCR of three positive samples collected in UTM performed with CFX96™ Real-Time PCR Detection System and relative Ct values for each sample. (B) Real-Time PCR of same three samples collected in UTM performed with SWM-01 Analyzer and relative Ct values for each sample.
The results obtained in this analysis suggest that both techniques are useful and experimentally reproducible for strongly positive samples since we recorded similar Ct values for each of them. Interestingly, two samples (13191 and 13153) analyzed with the SWM-01 instrument, are detected with a ‘delay’ of five and three cycles respectively, compared with the results obtained with the standard detection system. The reaction system of CFX96™ is 25 μL, which is about 2 times of SWM-01. Hence, the difference of sample capacity between two devices may result in the gap of the Ct value with different fluorescent intensity.
3.2.2.2 Weak positive samples in UTM: From previous outcomes, we tried to investigate the detection sensitivity even for weakly positive samples. We report results from RT-PCR of five weakly positive samples obtained from nasopharyngeal swabs and collected in UTM and one synthetic positive control with both detection systems (Figure 4).
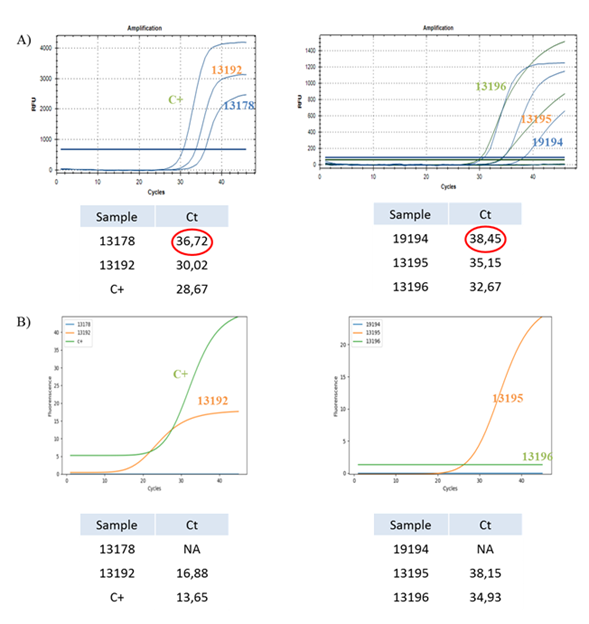
Figure 4: Sensitivity of different detection systems on weakly positive samples collected in UTM. (A) Real-Time PCR of four (13178, 19194, 13195 and 13196) weakly positive samples, one (13192) positive sample and one COVID-19 positive control performed with CFX96™ Real-Time PCR Detection System and Ct values for each sample. (B) Real-Time PCR of four (13178, 19194, 13195 and 13196) weakly positive samples, one (13192) positive sample and one COVID-19 positive control performed with SWM-01 Analyzer and Ct values for each sample.
We observed that positive samples (13192), including the positive control, are correctly detected with both detection systems and that the SWM-01 Analyzer can detect them earlier than the other instrument.
Otherwise, weakly positive samples are only partially detected. Samples with a low amount of viral RNA (13195 and 13196) are correctly detected with both instruments although the SWM-01 Analyzer detect them with a ‘delay’ of three cycles, while samples with a very low amount of viral RNA (13178 and 19194) are detected exclusively with a standard RT-PCR Detection System and not with the SWM-01 Analyzer. We assumed that the detection limit for the new instrument is about Ct<36.
In order to clarify the detection limits for samples with a limited amount of viral RNA, we performed a Real-Time PCR comparing results from both detection systems on three diluted samples (1:10 and 1:20 dilution) collected in UTM and on a positive control at same dilutions (Table 2).
|
Bio-rad Ct |
SWM-01 Ct |
||
|
Sample |
C+ |
27 |
26 |
|
13188 |
18 |
19 |
|
|
13192 |
27 |
28 |
|
|
Dilute sample 1:10 |
C+ |
30 |
27 |
|
13188 |
24 |
26 |
|
|
13192 |
33 |
NA |
|
|
Dilute sample 1:20 |
C+ |
36 |
32 |
|
13188 |
31 |
30 |
|
|
13192 |
NA |
NA |
Table 2: Summary of Ct values from Real-Time PCR of two positive samples in UTM and one positive control at initial concentration and at 1:10, 1:20 dilutions performed with both instruments.
Our data demonstrate that samples at 1:10 dilution are detected with both methods and results are comparable, conversely results obtained from samples at 1:20 dilution are not experimentally reproducible. The synthetic positive control is detected with a delay of 9 and 6 cycles with both instruments and the positive samples collected in UTM are both detected with a substantial delay compared to the sample at initial concentration.
4. Conclusions
In the last year COVID-19 rapidly evolved in a global pandemic causing clinical and diagnostic difficulties in several affected countries. Among different analytical approaches proposed in literature for large-scale laboratory screening [8, 10-12], we described a SWM-01 PCR Nucleic Acids Analyzer as a useful instrument for qRT-PCR and sensitive SARS-CoV-2 detection in nasopharyngeal swabs. The SWM-01 Analyzer combines the “microfluidic chip" technology with the sensitivity of a single-channel fluorescence PCR, allowing the simultaneous viral detection from 3 to 9 samples in a single microchip with 3 channels. The time required for the analysis is 45 minutes and considering that a classic qRT-PCR usually takes from 2 to 3 hours and allows the analysis of a smaller number of samples, it could represent an advantage for those laboratories that manage a large number of swabs every day or to manage cases of urgency. Comparing results with a standard qRT-PCR we analyzed 20 samples derived from nasopharyngeal swabs and collected in UTM, including both negative and positive samples, and we defined a detection limit for the new instrument as Ct<36. Weakly positive samples, with a limited viral charge, are not correctly assigned with the SWM-01 PCR Nucleic Acids Analyzer. We supposed that the detection limits are affected by the instrument itself [16]. Table 3 summarizes the comparison between this two methods (Table 3).
|
Technology |
SHINEWAY SWM-01 Analyzer (SWM-01) |
Conventional Real-Time PCR Detection System (as CFX96™ ) |
|
Assay to results time (excluded RNA isolation) |
42 minutes |
122 minutes |
|
Number of max targets simultaneously |
1 |
5 |
|
Number of sample for run |
9 |
96 |
|
Sensitivity |
Ct<36 |
Ct<40 |
|
Point of care |
Possible |
No |
Table 3: Performance comparison between SWM-01 and conventional qPCR.
From the table, is evident that these two methods have different strengths, SWM-01 is more rapid but the number of targets and samples are minor that conventional qPCR. On the other hand SWM-01 may find application in point of care because is easy used and moved. Finally, SWM-01 seems less sensitive that qPCR.
Weakly positive sample can cause the false-negative result due to the low concentration of targeted nucleic acid. During the PCR process, the fluorescent signal generated from amplification of positive sample should reach the detection limit so that the device can make correct determination.
Hence, positive sample in low concentration reduce the fluorescence intensity of reaction product at each cycle, which may result in uncertain detection outcome for PCR assay. On the one hand, to enhance the detection sensitivity, we could increase the sample capacity of the microchamber so that higher amount of viral DNA could be loaded. Instead, we could enhance the detection limit of the device by optimizing the fluorescent collection module and image analysis software.
Highly positive samples are correctly assigned with the innovative method and also with higher sensitivity. Moreover, since the global pandemic has left laboratories in difficulty due to the availability of diagnostic products, we also provide a new qRT-PCR protocol with the use of a smaller amount of reagents useful to avoid waste of material in the PCR reaction. This methods may be useful in case of urgent case or in specific laboratory condition, as samples that must be rerun in a short time or in a point of care.
In conclusion we can propose the SWM-01 PCR Nucleic Acids Analyzer as an interesting diagnostic tool suitable for routine diagnosis and for easier and faster laboratory screening.
Author Contributions
Conceptualization, Francesca Dragoni, Cristina Cereda and Stella Gagliardi; Data curation, Francesca Dragoni, Rosa Trotti, Cristina Cereda and Stella Gagliardi; Formal analysis, Francesca Dragoni, Cristina Cereda and Stella Gagliardi; Funding acquisition, Cristina Cereda; Investigation, Francesca Dragoni, Maria Garofalo, Yiteng Liu and Stella Gagliardi; Methodology, Francesca Dragoni, Maria Garofalo, Rosa Trotti, Cristina Cereda and Stella Gagliardi; Project administration, Cristina Cereda and Stella Gagliardi; Resources, Yiteng Liu, Cristina Cereda and Stella Gagliardi; Software, Francesca Dragoni and Stella Gagliardi; Supervision, Cristina Cereda; Validation, Francesca Dragoni, Maria Garofalo and Stella Gagliardi; Visualization, Francesca Dragoni, Maria Garofalo, Yiteng Liu and Stella Gagliardi; Writing – original draft, Francesca Dragoni, Yiteng Liu, Cristina Cereda and Stella Gagliardi; Writing – review & editing, Francesca Dragoni, Cristina Cereda and Stella Gagliardi.
Funding
Project was funded by the Italian Ministry of Health (Ricerca Corrente 2020).
Institutional Review Board Statement
The study was developed on existing samples collected during standard diagnostic tests (DGR regionale 3131/2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data is contained within the article.
Acknowledgments
We would like to thank Dr. Antonio Traversi, Dr. Yaying Hong and Prysmian Group for the support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li X, Geng M, Peng Y, et al. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal 10 (2020): 102-108.
- Zhang Z, Wang X, Wei X, et al. Multiplex quantitative detection of SARS-CoV-2 specific IgG and IgM antibodies based on DNA-assisted nanopore sensing. Biosens Bioelectron 181 (2021): 113134.
- Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol 41 (2020): 1100-1115.
- Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol 287 (2005): 1-30.
- Giri B, Pandey S, Shrestha R, et al. Review of analytical performance of COVID-19 detection methods. Anal Bioanal Chem 413 (2021): 35-48.
- Su S, Wong G, Shi W, et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol 24 (2016): 490-502.
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395 (2020): 497-506.
- Alpdagtas S, Ilhan E, Uysal E, et al. Evaluation of current diagnostic methods for COVID-19. APL Bioeng 4 (2020): 041506.
- Rodriguez C, de Prost N, Fourati S, et al. Viral genomic, metagenomic and human transcriptomic characterization and prediction of the clinical forms of COVID-19. PLoS Pathog 17 (2021): e1009416.
- Da Silva SJR, Silva CTAD, Guarines KM, et al. Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19. ACS Infect Dis 6 (2020): 2319-2336.
- Afzal A. Molecular diagnostic technologies for COVID-19: Limitations and challenges. J Adv Res 26 (2020): 149-159.
- Sreepadmanabh M, Sahu AK, Chande A. COVID-19: Advances in diagnostic tools, treatment strategies, and vaccine development. J Biosci 45 (2020): 148.
- Wang H, Liu Q, Hu J, et al. Nasopharyngeal Swabs Are More Sensitive Than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load. Front Med (Lausanne) 7 (2020): 334.
- Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn 20 (2020): 453-454.
- Chau CH, Strope JD, Figg WD. COVID-19 Clinical Diagnostics and Testing Technology. Pharmacotherapy 40 (2020): 857-868.
- Ping C, Sicen W. Application of microfluidic chip technology in pharmaceutical analysis: A review. Journal of Pharmaceutical Analysis 9 (2019): 238-247.
