Bioinformatic Analysis of Transcriptional Regulation by Nur77 in Central Nervous System and Immune System
Article Information
Fernando Faunes 1, María Estela Andrés2, Montserrat Olivares-Costa2,3*
1Departamento de Ciencias Biológicas, Facultad de Ciencias de la Vida, Universidad Andres Bello, Viña del Mar, Chile.
2Departmento de Biología Celular y Molecular, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile
3Departamento de Ciencias Biomédicas, Facultad de Medicina, Universidad Católica del norte.
*Corresponding author: Montserrat Olivares-Costa. Departamento de Biología Celular y Molecular, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile. Departamento de ciencias biomédicas, Facultad de Medicina, Universidad Católica del norte
Received: 22 December 2022; Accepted: 29 December 2022; Published: 12 July 2023
Citation: Faunes F, Andrés ME, Olivares-Costa M. Bioinformatic analysis of transcriptional regulation by Nur77 in central nervous system and immune system. Journal of Bioinformatics and Systems Biology. 6 (2023): 134-151.
Share at FacebookAbstract
Objective: The objectives of this work were to find genes regulated by Nur77 in neurons and to evaluate the possible common role of this transcription factor in neurons and lymphatic cells using published experimentally generated databases of ChIP-Seq and a microarray. We also characterized Nur77 binding throughout the genome.
Results: We identified 113 Nur77 target genes in neuronal stem cells and 116 in neuronal cells. Cell adhesion and anchoring processes emerged as regulated by Nur77 in neurons and lymphatic cells. We found 9 common genes regulated by Nur77. Finally, we described a significant distribution of Nur77 binding sites in strong enhancers and active promoters. This work is a first step to understand the role of Nur77 and its common targets in neurons and immune cells.
Keywords
Transcription factor, Gene expression, Nur77, Databases.
Transcription factor articles; Gene expression articles; Nur77 articles; Databases articles.
Transcription factor articles Transcription factor Research articles Transcription factor review articles Transcription factor PubMed articles Transcription factor PubMed Central articles Transcription factor 2023 articles Transcription factor 2024 articles Transcription factor Scopus articles Transcription factor impact factor journals Transcription factor Scopus journals Transcription factor PubMed journals Transcription factor medical journals Transcription factor free journals Transcription factor best journals Transcription factor top journals Transcription factor free medical journals Transcription factor famous journals Transcription factor Google Scholar indexed journals Gene expression articles Gene expression Research articles Gene expression review articles Gene expression PubMed articles Gene expression PubMed Central articles Gene expression 2023 articles Gene expression 2024 articles Gene expression Scopus articles Gene expression impact factor journals Gene expression Scopus journals Gene expression PubMed journals Gene expression medical journals Gene expression free journals Gene expression best journals Gene expression top journals Gene expression free medical journals Gene expression famous journals Gene expression Google Scholar indexed journals Nur77 articles Nur77 Research articles Nur77 review articles Nur77 PubMed articles Nur77 PubMed Central articles Nur77 2023 articles Nur77 2024 articles Nur77 Scopus articles Nur77 impact factor journals Nur77 Scopus journals Nur77 PubMed journals Nur77 medical journals Nur77 free journals Nur77 best journals Nur77 top journals Nur77 free medical journals Nur77 famous journals Nur77 Google Scholar indexed journals Databases articles Databases Research articles Databases review articles Databases PubMed articles Databases PubMed Central articles Databases 2023 articles Databases 2024 articles Databases Scopus articles Databases impact factor journals Databases Scopus journals Databases PubMed journals Databases medical journals Databases free journals Databases best journals Databases top journals Databases free medical journals Databases famous journals Databases Google Scholar indexed journals Central Nervous System articles Central Nervous System Research articles Central Nervous System review articles Central Nervous System PubMed articles Central Nervous System PubMed Central articles Central Nervous System 2023 articles Central Nervous System 2024 articles Central Nervous System Scopus articles Central Nervous System impact factor journals Central Nervous System Scopus journals Central Nervous System PubMed journals Central Nervous System medical journals Central Nervous System free journals Central Nervous System best journals Central Nervous System top journals Central Nervous System free medical journals Central Nervous System famous journals Central Nervous System Google Scholar indexed journals Gene Expression Omnibus articles Gene Expression Omnibus Research articles Gene Expression Omnibus review articles Gene Expression Omnibus PubMed articles Gene Expression Omnibus PubMed Central articles Gene Expression Omnibus 2023 articles Gene Expression Omnibus 2024 articles Gene Expression Omnibus Scopus articles Gene Expression Omnibus impact factor journals Gene Expression Omnibus Scopus journals Gene Expression Omnibus PubMed journals Gene Expression Omnibus medical journals Gene Expression Omnibus free journals Gene Expression Omnibus best journals Gene Expression Omnibus top journals Gene Expression Omnibus free medical journals Gene Expression Omnibus famous journals Gene Expression Omnibus Google Scholar indexed journals ChIP-Seq articles ChIP-Seq Research articles ChIP-Seq review articles ChIP-Seq PubMed articles ChIP-Seq PubMed Central articles ChIP-Seq 2023 articles ChIP-Seq 2024 articles ChIP-Seq Scopus articles ChIP-Seq impact factor journals ChIP-Seq Scopus journals ChIP-Seq PubMed journals ChIP-Seq medical journals ChIP-Seq free journals ChIP-Seq best journals ChIP-Seq top journals ChIP-Seq free medical journals ChIP-Seq famous journals ChIP-Seq Google Scholar indexed journals genome articles genome Research articles genome review articles genome PubMed articles genome PubMed Central articles genome 2023 articles genome 2024 articles genome Scopus articles genome impact factor journals genome Scopus journals genome PubMed journals genome medical journals genome free journals genome best journals genome top journals genome free medical journals genome famous journals genome Google Scholar indexed journals transcription articles transcription Research articles transcription review articles transcription PubMed articles transcription PubMed Central articles transcription 2023 articles transcription 2024 articles transcription Scopus articles transcription impact factor journals transcription Scopus journals transcription PubMed journals transcription medical journals transcription free journals transcription best journals transcription top journals transcription free medical journals transcription famous journals transcription Google Scholar indexed journals
Article Details
1. Introduction
Nur77 (also known as NGFI-B, TR3, and NR4A1) [1-3], is a transcription factor encoded by an immediate-early gene, and an orphan member of the nuclear receptor superfamily [4-5]. Nur77 has been largely studied in the immune system for its function inducing apoptosis of auto-reactive immature lymphocytes T and modulating the inflammatory response [6-7]. In the Central Nervous System (CNS), Nur77 is widely expressed, particularly in brain nuclei that receive dopaminergic and noradrenergic neurotransmission, which regulates the expression of Nur77 [8]. Brain pathologies characterized by an imbalance of catecholamine neurotransmission, such as anxiety, addiction, schizophrenia, and Parkinson disease are associated with changes in the expression of Nur77 [9-11]. Despite the growing amount of information about Nur77 in CNS, the lack of knowledge of Nur77 target genes in neurons hinders the study of its function. Here we analyzed public and experimentally generated databases to provide a list of Nur77-directly regulated genes in neurons.
Given that Nur77 is involved in the response to synaptic stimulation and control of metabolism in immune and nervous cells [12-15], we asked if Nur77 regulates the expression of a set of common genes in the nervous and immune systems. Our analysis revealed a group of genes that binds Nur77 on their promoters and cell adhesion and anchoring functions emerged as regulated by Nur77 in both cell types. We also characterized Nur77 binding sites throughout the genome, finding a significant distribution in strong enhancers and active promoters, reinforcing the function of Nur77 as a transcriptional activator. The work presented here is an approach pretending to guide the experimental focus regarding Nur77 investigation, solving in part the problem of lack of knowledge of Nur77 target genes and presenting new functions that can be attributed to this transcription factor in both the immune and nervous systems.
2. Methods
2.1 Data acquisition
We downloaded the ChIP-Seq peaks from Gene Expression Omnibus (GEO) corresponding to EGFP-Nur77 in K562 (GSE31363) [16] and endogenous Nur77 in NSC (GSM1603270) and NC (GSM1603273) [17]. The microarray data was obtained from the work published by Chen et al. in 2014, (GEO Accession; GSE76805) [12]. We used ENSEMBL gene annotation for human (hg19) and mouse (mm10) which were directly extracted as TxDb objects though GenomicFeatures and TxDb. Mmusculus.UCSC.mm10.ensGene R packages. Additional genomic annotations of different chromatin states and functional regions were extracted from UCSC Table browser, including the K562 Genome Segmentation by ChromHMM and Ensembl Regulatory Build.
2.2 Nur77 binding site characterization
The overlap between the different ChIP-Seq peaks and genomic features were calculated with the GenomicRanges R package using the function findOverlaps.
We intersected the genomic coordinates with K562 Genome Segmentation by ChromHMM and calculated the overlapping enrichment, by normalizing by total coverage of each chromatin state across the genome.
To obtain Nur77 target genes, we used the annotation of TSS and proximal promoters provided by the Ensembl Regulatory build for human (hg19) and mouse (mm10). The annotated TSS and proximal promoters were assigned to transcripts ID from Ensembl annotation if they were located within 2000 nt upstream and 500 nt downstream from a transcript start coordinate. Then, the function findOverlaps from Genomic Ranges was used to find Nur77 ChIP-Seq peaks that overlapped with TSS or proximal promoters from the Ensembl Regulatory build. We used the biomaRt R package to find the common gene symbol associated with each of the Ensembl IDs that were assigned to each TSS and proximal promoter that overlapped with a Nur77 ChIP-Seq peak.
2.3 Microarray analyses
We computed the differentially expression results from the microarray data published by Chen et al. in 2014 [12], to filter genes that have an adjusted p-value lower than 0.05 and an absolute value of log2 fold change greater than 1. We plotted the results using ggplot2 and highlighting the names of genes for which a Nur77 peak were detected in their promoters.
2.4 Ontology analysis
We use the Gene Ontology Consortium server (www.geneontology.org) [18-19] and processed using default parameters with PANTHER analysis tool [20].
3. Results and Discussion
3.1 Nur77 target genes in neurons:
We considered as Nur77 target genes, those genes that bind Nur77 in a window of - 2kb to + 500 bp from TSS in their promoters, according to our re-analysis of ChIP-Seq data of endogenous Nur77 of mouse neural stem cells (NSC) and NSC differentiated to neurons (NC) [17]. To select genes in which the binding of Nur77 influences transcription, we used a microarray of mRNA from mouse hippocampal pyramidal neurons overexpressing Nur77 [12] and selected genes with a significant change of expression after Nur77 overexpression (Figure. 1A). We identified 113 Nur77 target genes in NSC (Table S.1) and 16 out these genes changed their expression with a fold-change ≥ 2 after Nur77 overexpression (Figure.1B). In NC, we identified 116 Nur77 target genes (Table S.2) and 17 out of these genes changed their expression with a fold-change ≥ 2 after Nur77 overexpression (Figure. 1C). We found 53 Nur77 target genes common to NSC and NC, suggesting that these genes maintain Nur77 binding to their promoter during neuronal differentiation (Table S.3).
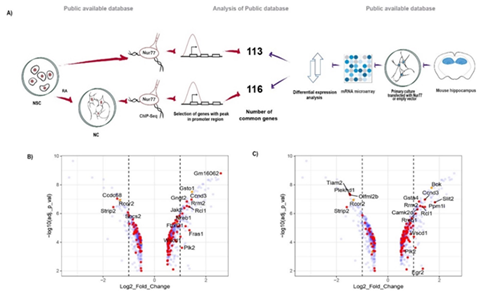
Figure 1: Nur77 target genes in neurons. A) Pipeline of the experimental procedure of public databases (12,17) and the analysis to select Nur77 target genes. B,C) Volcano plots representing genes that change their expression when Nur77 is overexpressed in pyramidal neurons (blue), genes that also have a peak.
of binding of Nur77 in their promoter (red dots) and TSS (orange dots) according to the ChIP-Seq form NSC (B), and from NC (C). Positive numbers indicate overexpression and negative numbers down-regulation. The dotted lines demarcate Log2 Fold of Change = ±1.
3.2 Binding of Nur77 on promoters of genes in the immune system and the central nervous system.
To find out whether Nur77 exerts a similar function in the nervous and immune systems and its conservation in human and mouse, we compared the binding peaks from two high quality experimentally generated ChIP-Seq databases: 1.- ChIP-Seq from the ENCODE project of overexpressing EGFP-Nur77 in the human chronic myelogenous leukemia cell line K562 (16) and 2.- ChIP-Seq of mouse NC (17) (Figure. 2A). Despite the differences between these two ChIP-Seq protocols, we found 271 genes with Nur77 common binding on their promoter (-2kb to +500bp from the TSS) in immune and neuronal cells (Figure. 2A, Table S.4).
GO analysis of these 271 genes (Figure. 2B and Table S.5) showed a significant enrichment of Nur77 target genes in ribonucleoprotein complex binding (fold of enrichment 5.2), cadherin bindings (fold of enrichment 3.94), cell adhesion molecule binding (fold of enrichment 3.05) and protein domain specific binding (fold of enrichment 2.66) (Figure. 2B). In the cellular component classification, Nur77 target genes were mainly enriched in categories of adhesion and junction: focal adhesion (fold of enrichment 3.45), cell-substrate adherent junction (fold of enrichment 3.43), cell-substrate junction (fold of enrichment 3.39), adherens junction (fold of enrichment 3.02), and anchoring junction (fold of enrichment 2.93). Two categories of nuclear localization were enriched: nuclear speck (fold of enrichment 3.33) and nuclear body (fold of enrichment 2.86) (Figure. 2B). Many proteins encoded by Nur77 target genes, which are common to the nervous and immune system, are ribonucleoproteins and adhesion molecules. This fact was strengthened by the enrichment of Nur77 target genes in nuclear bodies and areas of adherents and anchoring junctions (Figure. 2C).
In the biological process classification, Nur77 target genes were enriched in regulation of Endoplasmic-Reticulum-Associated protein Degradation (ERAD) pathway (fold of enrichment 12.91) and regulation of response to endoplasmic reticulum stress (fold of enrichment 8.01). Three GO terms related to interleukin signaling were enriched: interleukin-12-mediated signaling pathway (fold of enrichment 10.11) cellular response to interleukin-12 (fold of enrichment 9.68) and response to interleukin-12 (fold of enrichment 9.49). Nur77 target genes were also enriched in the regulation of protein autophosphorylation (fold of enrichment 9.49) and cell aging (8.22) GO terms (Figure. 2B).
Interestingly, 9 out of the 271 genes were also found in the set of genes that significantly changed their expression after Nur77 overexpression in neurons: AGAP3, BIRC5, DYM, ITGB3, KIF21B, MORN5, RREB1, STRIP2 and WEE1, suggesting that these genes are also regulated in the immune system. Previous evidence supports that Nur77 controls the expression of BIRC5 gene (21), validating our results. Further studies are required to fully validate Nur77 control over these genes, both in the nervous and immune system.
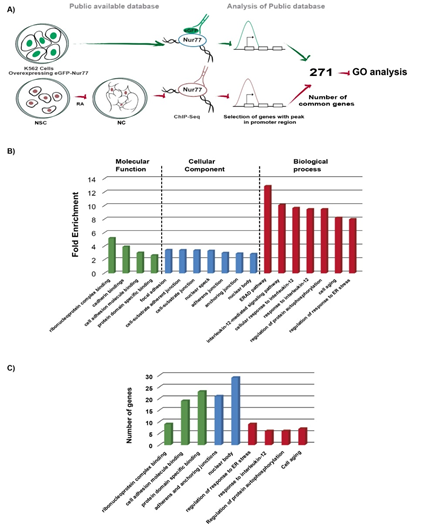
Figure 2: GO analysis of common target genes for Nur77 among the immune and nervous systems A) Pipeline of experimental procedure of public databases, and corresponding GO analysis. B) Enriched GO terms (more than 2.5 fold of change). C) The number of genes classified in each GO terms category. Molecular function classification in green bars, cellular component classification in blue bars and biological process classification in red bars. Cell adhesion molecule binding category includes cadherin binding GO term genes. Adherens and anchoring junctions category includes genes of adherens junction, anchoring junction, focal adhesion, cell-substrate adherent junction, and cell-substrate junction GO terms. Nuclear body category includes genes of nuclear speck GO term. Regulation of response to endoplasmic reticulum stress includes genes of regulation of ERAD pathway GO term. Response to interleukin-12 category includes genes of interleukin-12-mediated signaling pathways, cellular response to interleukin-12 and response to interleukin-12 GO terms (Table S.5).
3.3 Characterization of Nur77 binding sites throughout the genome:
To further characterize the binding profile of EGFP-Nur77, we compared the enrichment of ChIP-Seq peaks across 15 chromatin states defined by Ernest et al. for K562 cell line with the peaks of Nur77 [22]. We calculated the overlap enrichment across the EGFP-Nur77 peaks and chromatin states for K562 cell line, obtaining a significant enrichment of Nur77 peaks across chromatin states associated with transcription (Figure. 3A). The analysis of the coordinates of EGFP- Nur77 peaks, reported by ENCODE, with respect to the nearest transcription start site (TSS), revealed that Most of the Nur77 peaks are in a window of +1000 and -1000 bp from the nearest TSS (Figure. 3B). In this analysis, we used -2kb to + 500 bp range as a conservative range to increase coverage in the promoter area (upstream the TSS) while restricting the downstream coverage. It would be interesting to study different windows in the future to address a possible role of Nur77 when binding in downstream regions.
An enrichment analysis using the human chromatin segmentation model generated by Ernst, (2011) showed significant enrichment of Nur77 peaks in chromatin states associated with transcriptional regulatory regions, particularly in strong enhancers and active promoters (Figure. 3C). Two of chromatin states are described as strong enhancers (states 4 and 5), which differentiate in the occurrence of specific chromatin marks and distance to the TSS. Strong enhancer state number 4 presented a higher occurrence of histone 3 lysine 4 trimethylation (H3K4me3) and histone 3 lysine 9 acetylation (H3K9ac) and was closer to TSS than Strong enhancer state number 5. Nur77 was enriched in both chromatin states described as strong enhancers, exhibiting a log2 enrichment greater than 4 in chromatin state 4 (Figure. 3C). High enrichment of Nur77 binding was also observed in transcriptional transition states of chromatin. These areas presented similar characteristics to transcriptional elongation areas, but with an increased presence of H4K20me1 and H3K4me1 and more sensitive to DNAse [22], suggesting an intermediate state between the promoter activation and effective elongation. In contrast, we found a negative enrichment for Nu77 binding in transcriptional elongation areas. Nur77 was poorly enriched in states numbers 6 and 7, both described as weak enhancers which differ in the occurrence of H3K4me2 and DNase sensitivity [22]. Finally, negative enrichment was observed in heterochromatin regions, indicating the absence of Nur77 in inactive areas of the chromatin (Figure. 3C).
Altogether these data indicate that Nur77 is mostly associated with active sites of chromatin, concordant with the role of Nur77 as a transcriptional activator, also confirmed by the high presence of Nur77 in the TSS. Our data also shows that Nur77 binds transcriptional transition areas, suggesting that Nur77 is present in the promoters of its target genes independently of their state (active or inactive). On the other hand, the data suggests that the presence of Nur77 in enhancers would be limited to the active state.
In conclusion, our data analyses show that Nur77 is bound to the promoter of its target genes independently of their transcriptional state (weak, poised or active). In addition, our data suggests that the presence of Nur77 in enhancers is limited to the active state. We propose that Nur77 is always present in promoters but only binds to enhancers when it is upregulated, modulating thus transcription in response to stimuli.
Our analysis showed a strong participation of Nur77 target genes in anchoring and adhesion functions, which is consistent with the previously described roles of Nur77 in the modulation of neurite growth in neurons [13], and in the immune response [14-15], both processes that require interaction between cells and with the extracellular matrix.
Finally, genes found in this work as common targets of Nur77 in the nervous and immune systems are new and undescribed targets of this transcription factor. The work presented here is an approach pretending to guide the experimental focus regarding Nur77 investigation, solving in part the problem of lack of knowledge about Nur77 target genes and presenting new functions that can be attributed to this transcription factor in both the immune and nervous systems.
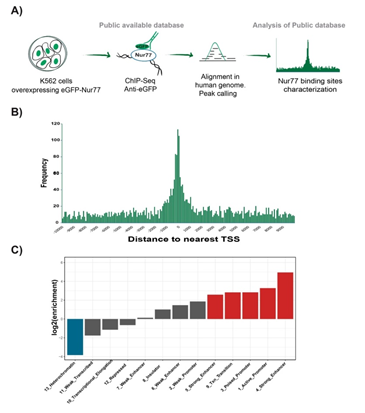
Figure 3: Nur77 binds to active promoters and strong enhancers in K562 cells. Analysis of publicly available database from the anti-EGFP ChIP-Seq experiment in K562 cell line overexpressing EGFP-Nur77 (16). A) Pipeline of experimental procedure of public database and bioinformatics analysis. B) Abundance of Nur77 binding respecting to the nearest TSS location. C) Enrichment of Nur77 binding sites across different chromatin regions defined by Ernst et al. in 2011 (22). In red, log2 of enrichment ≥ 2. In blue, log2 of enrichment 355 ≤ -2.
Limitations
The work presented here is a re-analysis of previously validated databases. However, differences in protocols or the overexpression of Nur77 could generate biases in the analyses. To be sure that genes described here are really modulated by Nur77, we were very restrictive in the selection process, this could lead to an underrepresentation of all genes regulated by Nur77 in neurons.
Abbreviations
ChIP-Seq: Chromatin immunoprecipitation followed by sequencing; CNS: Central nervous system; NC: Neuronal cells; NSC: Neuronal stem cells; GO: Gene ontology; TSS: Transcription start site.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Availability of data and materials
The datasets analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/. Access codes are detailed in the manuscript.
All data generated during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by Fondecyt grants 1150200 and 1191152. MO was supported by a Conicyt doctoral fellowship number 21140438 and by Fondo Postdoctorado Universidad Católica del Norte Nº 0003
Authors' contributions
MO, Design of the study, analysis and wrote the paper. FF, Discussed the results and wrote the paper. EA, Supervised the research, discussed the results, and wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Guillermo Parada for carrying out the bioinformatic analysis presented in this article.
References
- Hazel TG, Nathans D, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci USA 85 (1988): 8444-8.
- Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron 1(3) (1988): 183-188.
- Chawnshang C, Kokontis J, Shutsung L, et al. Isolation andcharacterization of human TR3 receptor: A member of steroid receptor superfamily. J Steroid Biochem 34 (1989): 391-395.
- Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: The second decade. Cell 83 (1995): 835-9.
- Flaig R, Greschik H, Peluso-Iltis C, et al. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem 280 (2005):19250-8.
- Woronicz JD, Calnan B, Ngo V, et al. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature 367 (1994): 277-81.
- McMorrow JP, Murphy EP. Inflammation: A role for NR4A orphan nuclear receptors. Biochemical Society Transactions 39 (2011): 688-693.
- Campos-Melo D, Galleguillos D, Sánchez N, et al. Nur transcription factors in stress and addiction. Frontiers in Molecular Neuroscience 6 (2013): 44.
- Xing G, Zhang L, Russell S, et al. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res 84 (2006): 36-56.
- Lévesque D, Rouillard C. Nur77 and retinoid X receptors: crucial factors in dopamine-related neuroadaptation. Trends in Neurosciences. Trends Neurosci 30 (2007): 22-30.
- Rouillard C, Baillargeon J, Paquet B, et al. Genetic disruption of the nuclear receptor Nur77 (Nr4a1) in rat reduces dopamine cell loss and L-Dopa-induced dyskinesia in experimental Parkinson’s disease. Exp Neurol 304 (2018): 143-153.
- Chen Y, Wang Y, Ertürk A, et al. Activity- induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron 83 (2014): 431-443.
- Jeanneteau F, Barrére C, Vos M, et al. The stress-induced transcription factor NR4a1 adjusts mitochondrial function and synapse number in prefrontal cortex. J Neurosci 38 (2018): 1335-1350.
- Cunningham NR, Artim SC, Fornadel CM, et al. Immature CD4 + CD8 + Thymocytes and Mature T Cells Regulate Nur77 Distinctly in Response to TCR Stimulation. J Immunol 77 (2006): 6660-6.
- Moran AE, Holzapfel KL, Xing Y, et al. T cell receptor signal strength in Treg and iNKT cell development 300 demonstrated by a novel fluorescent reporter mouse. J Exp Med 208 (2011): 1279-89.
- Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489 (2012): 57-74.
- Terranova C, Narla ST, Lee YW, et al. Global developmental gene programing involves a nuclear form of 306 fibroblast growth factor receptor-1 (FGFR1). PLoS One 10 (2015): 0123380.
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. Nature Genetics 25 (2000): 25-29.
- Carbon S, Douglass E, Dunn N, et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47(2019): 337-339.
- Mi H, Muruganujan A, Ebert D, et al. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47 (2019): 419-426.
- Yu C, Cui S, Zong C, et al. The orphan nuclear receptor NR4A1 protects pancreatic β-cells from endoplasmic reticulum (ER) stress-mediated apoptosis. J Biol Chem 290 (2015): 20687-20699.
- Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473 (2011): 43-49.
- Hoffmann R. A wiki for the life sciences where authorship matters: Nature Genetics Nat Genet 40 (2008): 1047-51.
Supplementary Materials
Table S.1: Nur77 target genes in NSC. Genes that bind Nur77 in their promoter regions, according to the ChIP-Seq of NSC, whose expression is modified when Nur77 is overexpressed in pyramidal neurons. Columns correspond to gene name, gene description from wikigene database [23], Nur77 peak location according to the ChIP-Seq from NSC (GSM1603270) [17], Log2 of the fold of change and p-value adjusted according to differential expression analysis of microarray from pyramidal neurons overexpressing Nur77 (GSE76805) [12] and gene identifier in the Ensembl database.
Table S.2: Nur77 target genes in NC. Genes that bind Nur77 in their promoter regions, according to the ChIP-Seq of NC, whose expression is modified when Nur77 is overexpressed in pyramidal neurons. Columns correspond to gene name, gene description from wikigene database [23], Nur77 peak location according to the ChIP-Seq from NC (GSM1603273) [17], Log2 of the fold of change and p-value adjusted according to differential expression analysis of microarray from pyramidal neurons overexpressing Nur77 (GSE76805) [12] and gene identifier in the Ensembl database.
Table S.3: Genes binding Nur77 in NSC and NC
|
Table S.3: Genes binding Nur77 in NSC and NC |
||
|
Gene Symbol |
||
|
NSC only |
NC only |
Common genes |
|
Aif1l |
2700012I20Rik |
Actr3b |
|
Akap13 |
4833418N02Rik |
Ankrd37 |
|
Asb2 |
4930429F24Rik |
Asxl3 |
|
Bcar3 |
Agap3 |
B3gat2 |
|
Blvrb |
Ahcyl2 |
Ccnd3 |
|
Brpf1 |
B3gnt2 |
Cdh2 |
|
Ccdc138 |
Bbs9 |
Chchd7 |
|
Ccdc68 |
Birc5 |
Cog1 |
|
Ccdc71l |
Bok |
Dock1 |
|
Ccnjl |
Camk2d |
Dym |
|
Cd59a |
Ccnd1 |
Etl4 |
|
Chaf1b |
Cd24a |
Fam171b |
|
Coq10b |
Cd83 |
Fbxw9 |
|
Dll1 |
Cdk2ap1 |
Fkbp1a |
|
Dnajc12 |
Cdkn2c |
Fkbp1b |
|
Dusp14 |
Cenpe |
Foxn2 |
|
Farp2 |
Crim1 |
Homer3 |
|
Fars2 |
Dusp10 |
Hsdl2 |
|
Fbxl21 |
Dusp6 |
Inpp4b |
|
Fgf18 |
Egfr |
Itgb3 |
|
Fgfr1 |
Egr2 |
Jmjd1c |
|
Flcn |
Erlin1 |
Jun |
|
Fras1 |
Errfi1 |
Larp4 |
|
Gm16062 |
Fabp3 |
Lmo2 |
|
Gngt2 |
Fanci |
Map6 |
|
Gsto1 |
Fbln7 |
Map7d2 |
|
H2-Q7 |
Gsta4 |
Myo5a |
|
Id1 |
Hr |
Nmrk1 |
|
Ift80 |
Itga6 |
Nod1 |
|
Jak2 |
Kif21b |
Numa1 |
|
Klhl25 |
Klf15 |
Pcx |
|
Ldlr |
Lhx9 |
Perp |
|
Lrba |
Lrp11 |
Plk2 |
|
Lrrfip1 |
Mat2b |
Ptger3 |
|
Map3k1 |
Morn1 |
Pygb |
|
Meis1 |
Morn5 |
Rcl1 |
|
Ndrg1 |
Nceh1 |
Rcor2 |
|
Nek11 |
Nes |
Rpp14 |
|
Nudt9 |
Nt5dc3 |
Rreb1 |
|
P4ha2 |
Olfml2b |
Rrm2 |
|
Ppp1r1b |
P2ry1 |
Runx1t1 |
|
Raly |
Pcgf5 |
Sertad1 |
|
Rhou |
Pkig |
Sfmbt1 |
Table S.4: Nur77 target genes common to lymphatic and neuronal systems.
Nur77 target genes, common to the immune and neural systems. Only genes exhibiting Nur77 binding in their promoters are listed. This set of genes was used for GO analysis.
|
Table S.4: Lymphatic and neuronal common target genes |
|
Gene symbol |
|
Cnnm3 |
|
Sdccag8 |
|
Sde2 |
|
Phlda3 |
|
Cnst |
|
Yod1 |
|
Pfkfb2 |
|
Kif21b |
|
Tmco1 |
|
Fnbp4 |
|
Acbd5 |
|
Hspa5 |
|
Mrrf |
|
Morn5 |
|
Ptpa |
|
Ak8 |
|
Abl1 |
|
Frmd4a |
|
Golga2 |
|
Ralgds |
|
Tnks1bp1 |
|
Cry2 |
|
Trim44 |
|
Prpf6 |
|
Ptpra |
|
Slc20a1 |
|
Itch |
|
Pcif1 |
|
Serf2 |
|
Cstf1 |
|
Samd10 |
|
Stard9 |
|
Otud7b |
|
Rsrc1 |
|
Selenot |
|
Leprot |
|
Pum1 |
|
Laptm5 |
|
Cc2d1b |
|
Ipp |
|
Capzb |
|
Ptp4a2 |
|
Patj |
|
Nr0b2 |
|
Lsm10 |
|
Tesk1 |
|
Tmem53 |
|
Coa7 |
|
Wasf2 |
|
Stk40 |
|
Zbtb8os |
|
Atp6v0b |
|
Rbbp4 |
|
Nom1 |
|
Mob1b |
|
Ywhah |
|
Agap3 |
|
Fbxo42 |
|
Mxra8 |
|
Cenpa |
|
Ccng2 |
|
Cep104 |
|
Ube3c |
|
Depdc5 |
|
Sh3bp2 |
|
Yes1 |
|
Krit1 |
|
Dhrs3 |
|
Ubr4 |
|
Srp72 |
|
Rpl22 |
|
Nsun5 |
|
Baz1b |
|
Cops6 |
|
Dr1 |
|
Gpn3 |
|
P2rx4 |
|
Srsf9 |
|
Casp2 |
|
Strip2 |
|
Pptc7 |
|
Mblac1 |
|
Chek2 |
|
Pms2 |
|
Vps29 |
|
Cav1 |
|
Rad9b |
|
Gtf2i |
|
Bbc3 |
|
Ercc1 |
|
Bcl2l13 |
|
Necap1 |
|
Hnrnpf |
|
Klhl42 |
|
Leng8 |
|
Serbp1 |
|
Zxdc |
|
Zfp36 |
|
Ube2m |
|
Foxp1 |
|
Map2k7 |
|
Lrrc27 |
|
Snrpa1 |
|
Lamtor1 |
|
Ilk |
|
Wee1 |
|
Fam103a1 |
|
Armc5 |
|
Setd1a |
|
Eef2k |
|
Mical2 |
|
Rpl27a |
|
Orai3 |
|
Fam174b |
|
Ucp2 |
|
Rrp8 |
|
Tfdp1 |
|
Rab8a |
|
Nutf2 |
|
Dctd |
|
Ap1g1 |
|
St3gal2 |
|
Herpud1 |
|
Cdh5 |
|
Kbtbd3 |
|
Mtss1l |
|
Irf2bp2 |
|
Coa6 |
|
Pdp2 |
|
Dnaaf1 |
|
Slc12a4 |
|
Cmip |
|
Adcy7 |
|
Qtrt1 |
|
Ccdc12 |
|
Limd1 |
|
Ctsh |
|
Nt5e |
|
Abhd5 |
|
Pif1 |
|
Vill |
|
Tcaim |
|
Usp2 |
|
Elp6 |
|
Als2cl |
|
Msl2 |
|
Trip4 |
|
Mob3a |
|
Cdk4 |
|
Lims1 |
|
Cdc34 |
|
Ascc3 |
|
Hace1 |
|
Cited2 |
|
Wdr18 |
|
Ascc1 |
|
Bsg |
|
Izumo4 |
|
Nfic |
|
Chst3 |
|
Ccdc6 |
|
Vps54 |
|
Rack1 |
|
Dbnl |
|
Mat2b |
|
Fnip1 |
|
Hnrnph1 |
|
Larp1 |
|
Snord96a |
|
Snord95 |
|
Tmem256 |
|
Rai1 |
|
Sqstm1 |
|
Lrrc75a |
|
Tbx2 |
|
Shpk |
|
Igfbp4 |
|
Pfn1 |
|
Rnft1 |
|
Ube2g1 |
|
Eno3 |
|
Fam222b |
|
Srsf1 |
|
Pitpna |
|
Snord4a |
|
Nfe2l1 |
|
Snx11 |
|
Rpl23a |
|
Twistnb |
|
Laptm4a |
|
Gna13 |
|
Mrc2 |
|
Unk |
|
Sec14l1 |
|
Fn3k |
|
Engase |
|
Hexim1 |
|
Ubald2 |
|
Gdi2 |
|
Birc5 |
|
B3galnt2 |
|
H3f3b |
|
Prcd |
|
Rreb1 |
|
Tmc8 |
|
Vdac2 |
|
Nkiras1 |
|
Vcl |
|
Ndufb9 |
|
Cyc1 |
|
Adk |
|
Tbca |
|
Mblac2 |
|
Plec |
|
Col4a3bp |
|
Chrac1 |
|
Drosha |
|
Gnb1l |
|
Hoxc10 |
|
Tef |
|
Serpind1 |
|
Top3b |
|
Slc4a8 |
|
Yars2 |
|
Txnrd2 |
|
Thap7 |
|
Ubald1 |
|
Rtl10 |
|
Hsf2bp |
|
Cd320 |
|
Sod2 |
|
Wrb |
|
Pkmyt1 |
|
Luc7l |
|
Brd2 |
|
Morc3 |
|
Dnph1 |
|
Rrp1b |
|
Tfeb |
|
Safb |
|
Safb2 |
|
Rpp21 |
|
Etf1 |
|
Impa2 |
|
Seh1l |
|
Dctn4 |
|
Fth1 |
|
Yif1a |
|
Dym |
|
Cxxc5 |
|
Bscl2 |
|
Trmt112 |
|
Ubxn1 |
|
Aip |
|
Tmem109 |
|
Prdx5 |
|
Ankrd13d |
|
Neat1 |
|
Hspa9 |
|
Coq5 |
|
Slc30a10 |
|
Ptgir |
|
Btbd19 |
|
Akt2 |
|
Tanc1 |
|
Mettl13 |
|
Itgb3 |
|
Ten1 |
|
Gsdmd |
|
Hist1h2bn |
|
Rab4a |
Table S.5: GO terms enriched in the analysis of Nur77 target genes.
GO terms enriched in the GO analysis and Nur77 target genes classified in each term. One table for each of the three principals GO categories is shown in separate sheets.
|
Biological Process |
|||
|
GO category |
Mapped Ids |
Gene name |
|
|
Regulation of response to endoplasmic reticulum stress |
Regulation of ERAD pathway |
Herpud1 |
Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein |
|
Ubxn1 |
UBX domain-containing protein 1 |
||
|
Yod1 |
Ubiquitin thioesterase OTU1 |
||
|
Rnft1 |
E3 ubiquitin-protein ligase RNFT1 |
||
|
Cav1 |
Caveolin-1 |
||
|
Rack1 |
Receptor of activated protein C kinase 1 |
||
|
Hspa5 |
Endoplasmic reticulum chaperone BiP |
||
|
Nfe2l1 |
Endoplasmic reticulum membrane sensor NFE2L1 |
||
|
Bbc3 |
Bcl-2-binding component 3 |
||
|
Interleukin-12-mediated signaling pathway; Cellular response to interleukin-12; Response to interleukin-12 |
Aip |
AH receptor-interacting protein |
|
|
Snrpa1 |
U2 small nuclear ribonucleoprotein A' |
||
|
Sod2 |
Superoxide dismutase [Mn], mitochondrial |
||
|
Hnrnpf |
Heterogeneous nuclear ribonucleoprotein F |
||
|
Pitpna |
Phosphatidylinositol transfer protein alpha isoform |
||
|
Hspa9 |
Stress-70 protein, mitochondrial;HSPA9 |
||
|
Regulation of protein autophosphorylation |
Eef2k |
Eukaryotic elongation factor 2 kinase |
|
|
Tnks1bp1 |
182 kDa tankyrase-1-binding protein |
||
|
Tesk1 |
Dual specificity testis-specific protein kinase 1 |
||
|
Birc5 |
Baculoviral IAP repeat-containing protein 5 |
||
|
Mob1b |
MOB kinase activator 1B |
||
|
Cav1 |
Caveolin-1 |
||
|
Cell aging |
Cited2 |
Cbp/p300-interacting transactivator 2 |
|
|
Ilk |
Integrin-linked protein kinase |
||
|
Lims1 |
LIM and senescent cell antigen-like-containing domain protein 1 |
||
|
Tbx2 |
T-box transcription factor TBX2 |
||
|
Morc3 |
MORC family CW-type zinc finger protein 3 |
||
|
Chek2 |
Serine/threonine-protein kinase Chk2 |
||
|
Ercc1 |
DNA excision repair protein ERCC-1 |
||
