An Empirical Study of Transfer Learning for Colorectal Polyps Image Segmentation
Article Information
Zhuo Zhou1*, Lin Fang2*, Bo Liu3, Jun Huang4,5,6#
1Health Management Center, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510620, Guangdong, China
2Department of Continuing Education, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510060, Guangdong, China
3Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, China.
4Department of Colorectal Surgery, Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510655, Guangdong, China
5Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510655, Guangdong, China
6Guangdong Institute of Gastroenterology, Guangzhou, 510655, Guangdong, China
*Corresponding author: Jun Huang, Department of Colorectal Surgery, Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510655, Guangdong, China.
Received: 15 October 2022; Accepted: 27 October 2022; Published: 04 November 2022
Citation: Zhuo Zhou, Lin Fang, Bo Liu, Jun Huang. An Empirical Study of Transfer Learning for Colorectal Polyps Image Segmentation. Archives of Clinical and Biomedical Research 6 (2022): 912-915.
Share at FacebookAbstract
Deep learning methods for medical image segmentation typically rely on pretrained models developed for natural images. The tremendous success of transfer learning raises the question: what makes a pretrained model good for medical image segmentation? In this paper, we explore properties of pretrained models on medical image segmentation. We compare the model performance on a polyp segmentation dataset and find that both the choice of network architecture and pretraining dataset are critical to the model’s transferability, while larger network does not always result in superior transfer learning performance.
Keywords
Medical Image; Neural Network; Polyps Segmentation; Transfer Learning; Semantic Segmentation
Article Details
1. Introduction
With the rapid development of deep learning, transfer learning from natural images has become a de-facto standard for solving many medical image problems, such as image classification, object detection, and segmentation. The most common way to train a medical image model is to take an existing network architecture that designed for natural image dataset along with the pretrained model weights and fine-tune the model with medical image data. Fine-tuning models pretrained on large-scale natural image dataset such as ImageNet [1] has achieved impressive results on several medical image applications, including chest X-ray classification [2], brain MRI segmentation [3], breast ultrasound image segmentation [4], COVID-19 detection using lung CT image [5]. Despite the popularity of transfer learning in medical image analysis, there has not been a systematic study of which aspects of the pretraining model affects the model’s generalization ability on medical images. In this work, we systematically investigate how pretrained models is related to the performance of polyp segmentation in colonoscopy images. The primary contributions of this paper are:
- For models pretrained on the same dataset, we find monotonic relationship between the Cityscapes segmentation performance and the Kvasir-SEG segmentation performance. This finding suggests that the better segmentation network architecture leads to improvement on medical image segmentation task.
- For models with the same segmentation method, a more powerful backbone leads to better transfer learning performance.
- We observe that models pretrained on datasets that are similar to the target dataset transfer better. Pretraining datasets that have larger scale and diversity help the transfer learning performance.
- For models with the same network architecture, increasing the network depth does not necessarily improve the transfer learning performance.
2. Method
2.1 Datasets
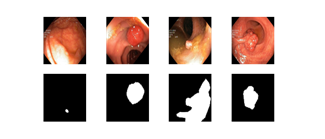
Our primary dataset, the Kvasir-SEG (6), contains 1000 gastrointestinal polyp images with varies resolution from 332 × 487 to 1920 × 1072 pixels. Each image has a corresponding polyps mask that was manually labeled and verified by an experienced gastroenterologist. These images are used to detect and assess polyps, which are precursors to colorectal cancer. As one of most common cancers in men and women, the colorectal cancer has a five-year survival rate of 10% when discovered in advanced stage, whereas when it is diagnosed in early stages, the five-year survival rate increased to 90% [7]. Such early diagnosis is achievable if polyps can be detected and removed before turning malignant. Therefore, having a reliable way to detect polyps is crucial for preventing and increasing the survival rate for colorectal cancer. Figure 1 depicts some example images from the Kvasir-SEG datasets, demonstrating the large variety of polyps in terms of shape, size, color, and texture. In this work, the polyp detection is formulated as a semantic segmentation problem, in which each pixel is classified either as polyps or background. For all models evaluated in this study, three metrics, Intersection-over-Union (IoU), Overall Accuracy (OA), Dice, are reported for the segmentation performance.
2.2 Experimental Setup
The process of initializing the neural network with weights that pretrained on a large-scale dataset, like ImageNet, and continuously fine-tuning on a target dataset is referred to as transfer learning. All the semantic segmentation networks used in our experiments use the MMSegmentation [8] implementation. For the Kvasir-SEG dataset, we split the data with an 80/20 ratio for training and test. If not mentioned otherwise, we use the following setup for all networks compared in our experiments. The Kvasir-SEG image is resized to 640×640 for training and test. During training, a 512×512 crop is randomly sampled from each image with the ImageNet per-pixel mean subtracted. We use the standard data augmentation by randomly flip the image with a ratio of 0.5. The standard color augmentation such as brightness, contrast, and saturation jitter as well as color normalization with ImageNet pixel mean and standard deviation are used to avoid overfitting. We fine-tune the network for 16,000 iterations with a batch size 8, which is equivalent to 160 epochs on the Kvasir-SEG dataset. We use SGD with an initial learning rate of 0.01. The weight decay is set to 0.005 and momentum is set to 0.9. We use the Polynomial decay teaching rate scheduler, where the learning rate of the current iteration equals to , with a power of 0.9. All the models in our experiments are trained with a single NVIDIA T4 GPU.
3. Results
To understand what affects the performance of transfer learning, we select multiple pretrained models and evaluate their performance by varying the following aspects: segmentation method, backbone of the segmentation network, pretrained dataset, the size of pretrained network, the length of pretraining, the learning rate and batch size of fine-tuning.
3.1 Effect of Segmentation Network Architecture
To examine the effect of segmentation methods, we select four pretrained models, FCN [9], PSP [10], DeepLabV3 [11], UPerNet [12], and fine-tune them on the Kvasir-SEG dataset. To isolate the segmentation network effect, we use the same backbone ResNet-18 (R18) [13] and all four networks are initialized with weights pretrained on the Cityscapes dataset [14]. Table 1 shows the model performance after fine-tuning on the Kvasir-SEG data, along with the pretrained model performance on the Cityscapes data. We find monotonic relationship between the Cityscapes pretraining performance and the transfer learning performance on Kvasir-SEG with Spearman ρ = 0.800 at p = 0.200. This finding suggests that better semantic segmentation network architectures transfer better on the Kvasir-SEG polyps segmentation task. Despite having a slightly lower mIoU than DeepLabV3 on the Cityscapes data, UPerNet achieved the best transfer learning performance on the Kvasir-SEG data. We attribute this to the relatively simple network architecture of UPerNet compared to DeepLabV3.
|
Method |
Pretr. model mIoU |
IoU |
OA |
Dice |
|
FCN |
Cityscapes 71.11 |
74.78 |
84.37 |
85.57 |
|
PSP |
Cityscapes 74.84 |
76.81 |
83.94 |
86.88 |
|
DeepLabV3 |
Cityscapes 76.70 |
79.15 |
85.87 |
88.36 |
|
UPerNet |
Cityscapes 76.02 |
79.88 |
86.21 |
88.82 |
Table 1: Performance of different segmentation networks on the Kvasir-SEG dataset.
3.2 Effect of Backbone for the Segmentation Network
We investigate whether stronger backbone leads to higher segmentation performance on the Kvasir-SEG data. We pick three representative backbones, including ResNet-50 (R50) [13], Vision Transformer base (ViT-B) [15], Swin Transformer base (Swin-B) [16], based on their image classification performance on the ImageNet dataset. We use the same segmentation method, UPerNet [12], and initialize the network with weights pretrained on the ADE20K [17] dataset. In this comparison, models with transformer backbone are trained using the AdamW [18] optimizer with the initial learning rate of 6E − 5. Segmentation performance of three models are displayed in Table 2. We find a perfect association of rank, with Spearman ρ = 1 at p = 0, between the backbone classification performance on ImageNet and the UPerNet segmentation performance on Kvasir-SEG. This finding suggests that within the same segmentation method, stronger backbone leads to better segmentation performance on medical image segmentation task.
|
Method |
Backbone ImageNet Acc. |
IoU |
OA |
Dice |
|
UPerNet |
R50 76.1 |
80.11 |
86.71 |
88.96 |
|
UPerNet |
ViT-B 77.9 |
82.88 |
88.44 |
90.64 |
|
UPerNet |
Swin-B 84.5 |
84.88 |
90.34 |
91.82 |
Table 2: Performance of UPerNet with different backbones on the Kvasir-SEG dataset.
3.3 Effect of Pretraining Dataset
We study the effect of pretraining dataset by fine-tuning the same model with four different initializations (pretrained weights) on the Kvasir-SEG. The same segmentation network, FCN with HRNet-48 backbone, pretrained on three different segmentation datasets, including Cityscapes [14], Pascal VOC 2012 [19], ADE20K [17], are used in this comparison. We also include an ImageNet pretrained model, where only the backbone is pretrained, as a baseline. The segmentation performance of the four models are reported in Table 3. As we can see that there is relatively large performance gap between the ImageNet pretrained model and the other three models that are pretrained on segmentation datasets. This is expected as the model pretrained on segmentation dataset has the entire network trained while the model pretrained on ImageNet only has the backbone trained but not the segmentation branch of the architecture. Among the three segmentation datasets, the model pretrained on ADE20 achieved the best performance on Kvasir-SEG, followed by Pascal VOC and Cityscapes. We suspect that the similarity between the target dataset and the pretraining dataset is what causes the difference in transfer learning performance. Cityscapes is an autonomous driving dataset contains driving scenes, which has a totally different visual appearance than the medical image. While Pascal VOC and ADE20K both cover daily scenes, ADE20K contains more training images (22,210 vs. 10,103) and a larger variety of classes (150 vs. 20). This finding suggests that pretraining dataset that is similar to the target dataset and has larger scale and more diversity helps improve the transfer learning performance.
|
Method |
Backbone |
Pretrained Dataset |
IoU |
OA |
Dice |
|
FCN |
HRNet-48 |
ImageNet |
77.79 |
85.17 |
87.51 |
|
FCN |
HRNet-48 |
Cityscapes |
81.76 |
87.4 |
89.96 |
|
FCN |
HRNet-48 |
Pascal VOC 2012 |
82.95 |
88.37 |
90.68 |
|
FCN |
HRNet-48 |
ADE20k |
83.76 |
89.06 |
91.16 |
Table 3: Performance of FCN with different pretrained weights on the Kvasir-SEG dataset.
3.4 Effect of the Size of Pretrained Model
We examine whether larger models perform better than smaller models on the Kvasir-SEG dataset, where the size of model is measured by number of layers. We compare the performance of ResNet with three different depths. Table 4 shows the segmentation performance of model with different depths. The model with ResNet-101 backbone achieved the best segmentation performance, while the model with ResNet-50 performed worse than the model with ResNet-18 backbone. We find no monotonic relationship between the network depth and the Kvasir-SEG segmentation performance.
|
Method |
Backbone |
Pretrained Dataset |
IoU |
OA |
Dice |
|
FCN |
R18 |
Cityscapes |
74.78 |
84.37 |
85.57 |
|
FCN |
R50 |
Cityscapes |
70.85 |
82.3 |
82.94 |
|
FCN |
R101 |
Cityscapes |
79.3 |
85.93 |
88.46 |
Table 4: Performance of FCN with different backbone depths on the Kvasir-SEG dataset.
4.Conclusion
In this work, we study the transfer learning performance of pretrained models on polyp segmentation. We show that both the segmentation method and backbone choice positively affect the transfer learning performance on the Kvasir-SEG dataset. We also show that a pretraining dataset that has higher similarity to the target dataset, larger scale and diversity helps transfer learning. Better pretraining performance is often provided by deeper networks, but this achievement does not always translate into transfer learning.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [Grant No. 81972885], the 1010 project of the 6th Affiliated Hospital of Sun Yat-sen University [1010CG (2020)-20].
References
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks, in: Advances in neural information processing systems (2012): 1097-1105.
- Ke A, Ellsworth W, Banerjee O, et al. Chextransfer: performance and parameter efficiency of imagenet models for chest x-ray interpretation, in: Proceedings of the Conference on Health, Inference, and Learning (2021): 116-124.
- Yamanakkanavar N, Choi JY, Lee B. Mri segmentation and classification of human brain using deep learning for diagnosis of alzheimer’s disease: a survey. Sensors 20 (2020): 3243.
- Xu Y, Wang Y, Yuan J, et al. Medical breast ultrasound image segmentation by machine learning. Ultrasonics 91 (2019): 1-9.
- Ahuja S, Panigrahi BK, Dey N, et al. Deep transfer learning-based automated detection of covid-19 from lung ct scan slices. Applied Intelligence 51 (2021): 571-585.
- Jha D, Smedsrud PH, Riegler MA, et al. Kvasir-seg: A segmented polyp dataset, in: International Conference on Multimedia Modeling, Springer (2020): 451-462.
- Bernal J, Tajkbaksh N, Sanchez FJ, et al. Comparative validation of polyp detection methods in video colonoscopy: results from the miccai 2015 endoscopic vision challenge. IEEE transactions on medical imaging 36 (2017): 1231-1249.
- Contributors M. MMSegmentation: Openmmlab semantic segmentation toolbox and benchmark (2020).
- Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition (2015): 3431-3440.
- Zhao H, Shi J, Qi X, et al. Pyramid scene parsing network, in: Proceedings of the IEEE conference on computer vision and pattern recognition (2017): 2881-2890.
- Chen LC, Papandreou G, Schroff F, et al. Rethinking atrous convolution for semantic image segmentation, arXiv preprint arXiv:1706.05587 (2017).
- Xiao T, Liu Y, Zhou B, et al. Unified perceptual parsing for scene understanding, in: Proceedings of the European conference on computer vision (ECCV) (2018): 418-434.
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition (2016): 770-778.
- Cordts M, Omran M, Ramos S, et al. The cityscapes dataset for semantic urban scene understanding, in: Proceedings of the IEEE conference on computer vision and pattern recognition (2016): 3213-3223.
- Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: Transformers for image recognition at scale, arXiv preprint arXiv: 2010.11929 (2020).
- Liu Z, Lin Y, Cao Y, et al. Swin transformer: Hierarchical vision transformer using shifted windows, in: Proceedings of the IEEE/CVF International Conference on Computer Vision (2021): 10012-10022.
- Zhou B, Zhao H, Puig X, et al. Scene parsing through ade20k dataset, in: Proceedings of the IEEE conference on computer vision and pattern recognition (2017): 633-641.
- Loshchilov I, Hutter F. Decoupled weight decay regularization, in: International Conference on Learning Representations (2019).
- Everingham M, Van Gool L, Williams CKI, et al. The PASCAL Visual Object Classes Challenge 2012 (VOC2012) Results (2012).