Activation of Human Dendritic Cells by Nanoparticles Carrying CTL Epitopes of Non-Structural Proteins of Hepatitis C Virus
Article Information
Kuprianov Victor1, 2, *, Lyudmila Nikolaeva2, Anna Zykova1, Anna Dedova2, Artemiy Vakhrameev2, Nikolai Ravin1
1Department of Molecular Biology of Microorganisms, Scryabin Institute of Bioengineering, Research Center of Biotechnology of the Russian Academy of Sciences, Moscow, Russian Federation
2Department of Molecular Virology and Antiviral Drugs, Ivanovsky Institute of Virology, Gamaleya National Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation, Moscow, Russian Federation
*Corresponding author: Victor Kuprianov, Federal Research Centre “Fundamentals of Biotechnology” of the Russian Academy of Sciences. Address: 33 Leninsky prospect, build 2, Moscow 119071, Russian Federation.
Received: 24 December 2021; Accepted: 04 January 2022; Published: 14 January 2022
Citation:
Kuprianov Victor, Lyudmila Nikolaeva, Anna Zykova, Anna Dedova, Artemiy Vakhrameev, Nikolai Ravin. Activation of human dendritic cells by nanoparticles carrying CTL epitopes of non-structural proteins of hepatitis C virus. Archives of Microbiology and Immunology 6 (2022): 39-50.
Share at FacebookAbstract
The aim of this study was to produce immunogenic nanoparticles carrying cytotoxic T lymphocyte (CTL) epitopes of hepatitis C virus non-structural proteins. We have obtained recombinant proteins forming virus-like particles and containing the sequences of self-assembling peptides (SAP), PADRE, and CTL epitopes. Using atomic force microscopy (AFM), the size of thus obtained nanoparticles was shown to be dependent on number of CTL epitopes located at the C terminus of recombinant proteins. Recombinant protein aggregating into virus-like particles (VLP) consisted of SAP and a helical linker. It formed 16-18 nm homogeneous particles, as shown by AFM. VPL-PE1, which differs from VPL by the presence of PADRE and CTL epitopes from NS3, forms 25-30 nm nanoparticles. The VLP-PE2 protein, different from VLP-PE1 by the presence of NS4a and NS4b CTL epitopes, formed 70-80 nm nanoparticles. The size of nanoparticles depended on the presence of SAP and the number of inserted epitopes. These nanoparticles activated human dendritic cells (DCs) that, in turn, stimulated autologous T lymphocytes. The proliferative activity and interferon-γ (IFN-γ) production by the stimulated T lymphocytes were evaluated by their secondary stimulation with commercially available mixture of peptides from non-structural proteins of hepatitis C virus. The greatest stimulating effect on T lymphocytes was exerted by DCs activated by nanoparticles, consisting of the recombinant mosaic protein with SAP at the N terminus and CTL epitopes of NS3, NS4a, and NS4b proteins at the C terminus. The presence of SAP in recombinant proteins’ sequences increased their immunogenicity.
Keywords
self-assembling peptides, nanoparticles, recombinant mosaic proteins, hepatitis C virus CTL epitopes, dendritic cells, T lymphocytes
self-assembling peptides articles, nanoparticles articles, recombinant mosaic proteins articles, hepatitis C virus CTL epitopes articles, dendritic cells articles, T lymphocytes articles
Article Details
1. Introduction
Hepatitis C virus (HCV) is an enveloped RNA virus belonging to the Flaviviridae family. HCV causes an acute infectious liver disease with a high risk of turning into chronic hepatitis C (CHC). In many countries, 1-2% of the population has CHC that may lead to the development of cirrhosis and hepatocellular carcinoma (HCC) [1, 2]. In 2016, WHO declared that hepatitis C may be eliminated by 2030 due to significant progress in the treatment of this disease with powerful direct-acting antivirals (DAAs). However, the risk of recurrent infection, HCC development, reinfection, and formation of occult hepatitis C remains even after successful treatment with DAAs [3-5]. Obviously, complete elimination of hepatitis C is impossible without developing an effective vaccine [6].
Modern scientific advances enable the development of a vaccine against hepatitis C, using various technological approaches. Thus, research is underway on the development of candidate vaccines based on recombinant proteins, synthetic peptides, virus-like particles, and viral vectors expressing various HCV antigens. For more than 30 years, scientists have been working to create a vaccine against hepatitis C, but so far there have been no radical successes [7]. However, the accumulated knowledge and new approaches give hope for the development of the vaccine [8, 9].
One of promising approaches to the creation of vaccines against hepatitis C is designing nanovaccines based on mosaic recombinant polypeptides that simultaneously contain immunogenic sequences of viral epitopes as well as adjuvant and signal sequences [10-12]. This approach was used in the present study. Cellular immune response to the epitopes of viral non-structural proteins NS3, NS4a, and NS4b has an essential role in the elimination of HCV in acute hepatitis C [13-15]. Therefore, in this study, immunogenic CTL epitopes from NS3, NS4a, and NS4b were included in a recombinant protein that also contained the PADRE sequence and the peptides that stimulated the assembly into nanoparticles. The PADRE (Pan HLA-DR-binding epitope) sequence was used as an internal adjuvant carrying a Th epitope [16]. Human dendritic cells (DCs) were activated by these nanoparticles. The aim of this study was to produce nanoparticles from a mosaic recombinant protein containing CTL epitopes from HCV proteins NS3, NS4a, and NS4b, adjuvant and signal sequences, and to analyze their effect on human DCs.
2. Materials and Methods
2.1 Bacteria and Plasmids
- coli DLT1270 and E. coli DH5a were used for cloning. E. coli DLT1270, transformed by a pQE30 plasmid containing the target protein sequence, was used for expression. The pQE30 plasmid was modified by inserting a glycine linker at the BamH1-Sac1 sites to improve the interaction of the hexahistidine site with the Ni sorbent during purification. Nucleotide sequences optimized for E. coli encoded the recombinant proteins.
2.2 Plasmids Construction
The nucleotide sequences encoding recombinant proteins were obtained by annealing the oligonucleotides synthesized by Synthol (Russia). The recombinant proteins consisted of the following fragments starting from the N terminus: the sequences of hexahistidine, self-assembling peptides (SAP), spiral linker (Sp), Th epitope (PADRE), and TCL epitopes of NS3, NS4a, and NS4b proteins belonging to HCV subtype 1b. These sequences were inserted into the expressing plasmid pQE30, using the Sac1 and Ecor5 restriction sites and the latter was additionally introduced after Sac1. Sequence selection to enhance supercoiling of amino acid chains (helices) was carried out using the program available at https://embnet.vital-it.ch/software/COILS_form.html.
2.3 Expression and Purification of Recombinant Proteins
Expression and purification of recombinant proteins were carried out as described earlier with minor modifications [12]. Artificial genes encoding SAP and multiepitope recombinant HCV proteins were cloned into the pQE30 plasmid. Recombinant proteins carrying N-terminal polyhistidine tags were expressed in E. coli DLT1270 cells. The target proteins were eluted with SPT buffer (10mM Na2HPO4, 10mM Tris-HCl, pH 8.0) containing 1M imidazole and 4.5M urea. The eluted proteins were then refolded by dialysis to form nanoparticles. The eluates were dialyzed (1:500 volume ratio) overnight against 1M urea in phosphate-buffered saline (PBS) (pH 7.3) at 8°C and against 0.5 M urea in PBS (pH 7.3) at 8°C. The samples were then dialyzed (1:500 volume ratio) twice overnight against PBS (pH 7.3) at 8°C.
The protein concentration was determined by the Bradford method, using the Bio-Rad protein assay kit (Bio-Rad, USA). Recombinant proteins were analyzed by SDS polyacrylamide gel (10%) electrophoresis, using the Laemmli method, with molecular markers ranging from 10 kDa to 250 kDa (BioRad, #161-0363).
2.4 Atomic force Microscopy Imaging (AFM)
AFM images of the purified recombinant proteins were obtained using an NTEGRA Prima (NT-MDT, Russia) system with silicon cantilevers with Au reflective coating and a tip with a curvature radius of ~35 (nm) (NSG01, NT-MDT, Russia). After refolding, recombinant proteins were diluted with PBS, deposited onto freshly cleaved mica, and dried at room temperature. PBS was used as a negative control.
2.5 DCs Generation and Activation
The primary culture of human monocytes was obtained from peripheral blood leukocytes isolated by centrifugation in Ficoll-Hypaque density gradient and subsequent purification on magnetic beads with positive selection of CD14+ cells (MACS, Miltenyi Biotec). To obtain DCs, monocytes were cultured in RPMI-1640 medium with 10% fetal bovine serum, stimulating factor GM-CSF (1000 U/ml), and interleukin 4 (500 U/ml) (SciStoreLab, Russia) for 5 days. To obtain mature DCs, they were further incubated with the addition of lipopolysaccharide (LPS, 1 µg/ml) from E. coli (Sigma-Aldrich, Germany) for 2 days [17]. Blood donors were three healthy volunteers. The study conforms to the ethical standards of the Declaration of Helsinki.
Mature DCs were incubated in RPMI-1640 medium with 2% autogenous human serum and 10µg/ml recombinant protein for 3 days. The DCs were then pre-treated with mitomycin C (30 µg/ml) for 40 min at 37°C and washed 4 times with PBS before co-cultivation with lymphocytes.
2.6 Analysis of the Proliferative Activity of T lymphocytes
T lymphocytes were isolated from human peripheral blood leukocytes. The lymphocytes were cryopreserved after monocytes were removed. The lymphocytes were thawed and purified using the Pan T Cell Isolation Kit II (Miltenyi Biotec, Germany). Purified T lymphocytes were co-cultured with the stimulated DCs at 10:1 ratio for 5 days. A medium containing a PepTivator mixture of peptides “HCV 1b NS3, NS4” (Miltenyi Biotec, Germany) was then added to concentration of 1µg/ml and incubated for 24 hours. As a positive control, cells were also stimulated in separate wells with 10µg/ml phythaemagglutinin (PHA, PanEco, Russia). The proliferative activity was evaluated using CCK-8 (Sigma-Aldrich, Germany). The stimulation index (SI) was measured as the ratio of optical density of stimulated cells to that of non-stimulated cells. Each measurement was repeated three times.
2.7 Determination of IFN-γ in the Culture Medium
For 3 days, the content of IFN-γ in the culture medium was determined using the Gamma-Interferon-ELISA-BEST test system (Vector Best, Russia). Each measurement was repeated three times.
2.8 Statistical Analysis
The quantitative data was calculated as mean ± 2SEM. A Student's t test was used to assess the significance of differences. P values <0.05 were taken as statistically significant.
3. Results
3.1 Design and expression of recombinant proteins containing HCV epitopes
Certain viral antigenic epitopes in the form of synthetic peptides can induce an immune response which, however, will be very weak due to the small size of the peptides. It is known that these peptides’ immunogenicity can be increased by attaching them to a large protein or virus-like particles or nanoparticles [18]. In this study, we have constructed nanoparticles based on self-assembling mosaic recombinant polypeptides reproducing HLA-A2-restricted CTL epitopes of non-structural proteins of HCV. Self-assembling peptides developed by Raman were used with a slight modification, which enhances supercoiling of recombinant proteins and improves assembly of nanoparticles that are similar in size to virus-like particles [19].
When designing mosaic recombinant proteins, we used the well-known immunogenic CTL epitopes of non-structural HCV proteins: NS3, amino acid (aa) sequences 1069-1082 and aa 1169-1177 [20-23]; NS4a, aa 1689-1711 [14, 15, 21, 24]; NS4b, aa 1763-1816 and aa 1851-1859 [22, 25, 26]. Dilysine linkers (KK) were inserted between epitopes to improve their processing and presentation [27]. To enhance immunogenicity, the PADRE sequence (AKFVAAWTLKAAA) was inserted [16]. The structure of the first protein designated as PE2 was His-PADRE-NS3 epitopes-NS4b epitopes-NS4a epitopes. Its amino acid sequence is shown below:
MRGSHHHHHHGSACELAKFVAAWTLKAAADKKFLATCINGVCWTVYKKLLCPSGHVVKKKKHMWNFISGVQYLAGLSTLPGNPAIASLMAFTASITSPLTTQYTLLFNILGGWVKKILAGYGAGVKKLSTWVLVGGVLAALAAYCLTTGSVVIVGRIVLSGKPAIIPDREVLYQEFDEMEECDI
The structure of the second protein designated as VLP was His-SAP-Sp. Its amino acid sequence is shown below:
MRGSHHHHHHGSACELDMELRELQETLAALQDVRELLRQQVKQITFLKCLLMGGRLLCRLEELERRLEELERRLEELERRDLEEAAEEKKEEAAEEKKEEAAEEKKEEAAEED.
The structure of the third protein designated as VLP-PE1 was His-SAP-Sp-PADRE-NS3 epitopes. The amino acid sequence is shown below:
MRGSHHHHHHGSACELDMELRELQETLAALQDVRELLRQQVKQITFLKCLLMGGRLLCRLEELERRLEELERRLEELERRDLEEAAEEKKEEAAEEKKEEAAEEKKEEAAEEDL AKFVAAWTLKAAAD KKFLATCINGVCWTVYKKLLCPSGHVVKK.
The fourth protein designated as VLP-PE2 had the following structure: His-SAP-Sp-PADRE-NS3 epitopes -NS4b epitopes -NS4a epitopes. The amino acid sequence is shown below:
MRGSHHHHHHGSACELDMELRELQETLAALQDVRELLRQQVKQITFLKCLLMGGRLLCRLEELERRLEELERRLEELERRDLEEAAEEKKEEAAEEKKEEAAEEKKEEAAEEDLAKFVAAWTLKAAADKKFLATCINGVCWTVYKKLLCPSGHVVKKKKHMWNFISGVQYLAGLSTLPGNPAIASLMAFTASITSPLTTQYTLLFNILGGWVKKILAGYGAGVKKLSTWVLVGGVLAALAAYCLTTGSVVIVGRIVLSGKPAIIPDREVLYQEFDEMEECDI
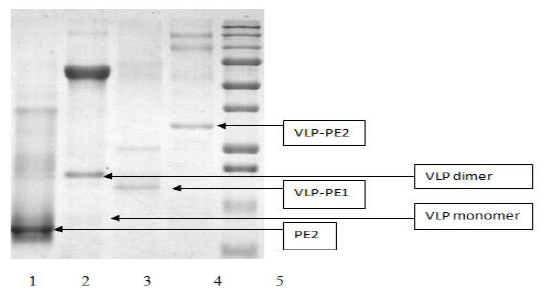
As all recombinant proteins were expressed in an insoluble form, they were dissolved in 7M guanidine hydrochloride and purified on the Ni sorbent (as described in Materials and Methods). The purified proteins were analyzed by polyacrylamide gel electrophoresis (Fig. 1).
3.2 Atomic force Microscopy Imaging of Recombinant Proteins
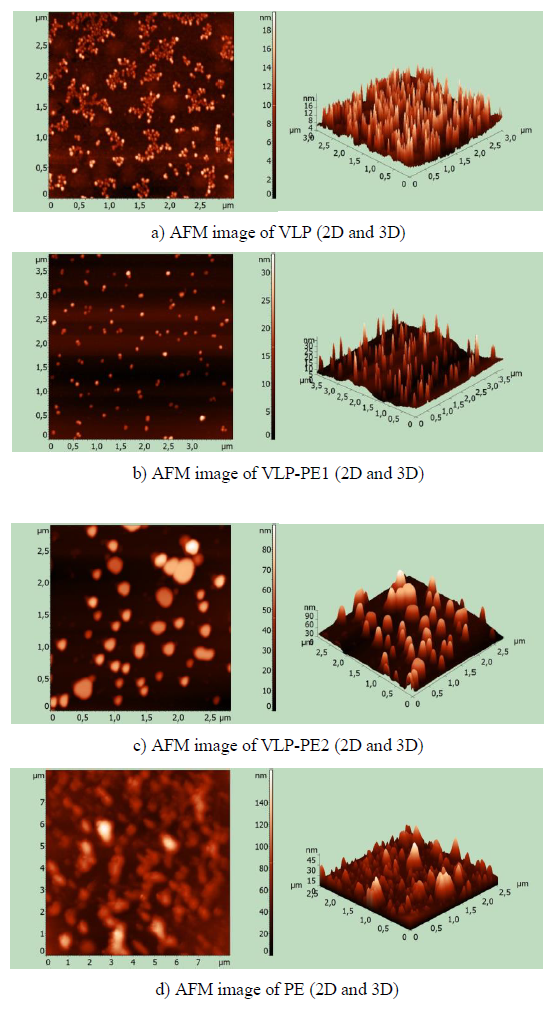
After refolding, the purified recombinant proteins were examined using atomic force microscopy (AFM). All recombinant proteins containing self-assembling peptides formed nanoparticles. Nanoparticles, containing a self-assembling peptide and a helical linker, formed homogeneous 16-18 nm particles (Fig. 2a). These particles formed aggregates. In the VLP-PE1 protein, which differs from VLP by the presence of PADRE and T cell epitopes from NS3, the nanoparticles were larger (25-30 nm) and less aggregated (Fig. 2b). The VLP-PE2 protein, which differed from VLP-PE1 by the presence of NS4a and NS4b, formed even larger nanoparticles 70-80 nm in size (Fig. 2c). The PE2 protein that contained no self-assembling peptides also formed aggregates, but more amorphous and inhomogeneous in size (from 10 to 150 nm) (Fig. 2d).
3.3 The Effects of the Recombinant Proteins on DCs
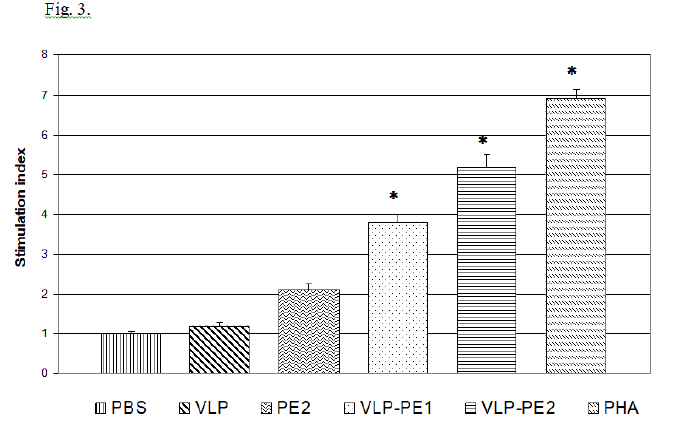
T lymphocytes stimulated with activated DCs become able to enhance proliferation during secondary stimulation with immunogenic peptides. The proliferative activity of lymphocytes stimulated with activated DCs is shown in Fig. 3.
It follows from the data presented in Fig. 3 that the presence of self-assembling peptides in a recombinant protein is associated with a more efficient activation of DCs and, accordingly, with a more pronounced proliferative activity of the stimulated T lymphocytes. The stimulation index (SI) value depended on the recombinant protein’s structure. Thus, SI of PE that does not contain amino acid sequence of self-assembling peptides comprised 2.1, which was not statistically significant, while VLP-PE1 and VLP-PE2 containing self-assembling peptides had higher SI values of 3.8 and 5.2, respectively (p <0.01). Thus, the presence of self-assembling peptides in the sequence of recombinant protein increases its immunogenicity.
3.4. Production of IFN-γ by T lymphocytes
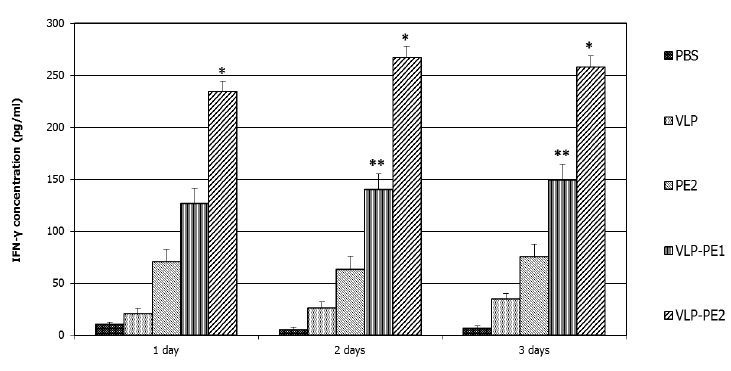
IFN-γ is involved in the regulation of nearly all phases of the immune and inflammatory responses, including the activation and differentiation of T, B, and NK cells and macrophages. IFN-γ secretion is characteristic of Th1 lymphocytes that secrete it only when activated. IFN-γ also increases the expression of class I MHC proteins on professional APCs and thus promotes antigen presentation to helper T cells. Therefore, it was important to analyze the impact of our recombinant proteins on the production of IFN-γ by lymphocytes. The accumulation of IFN-γ reflects the efficiency of stimulation of T lymphocytes by DCs presenting CTL epitopes. The greatest accumulation of IFN-γ was observed in the case of DCs activated by VLP-PE2 (267±8 pg/ml), with slightly less accumulation noted with DCs activated by VLP-PE1 (146±12 pg/ml) and very little accumulation in the case of DCs activation by PE2 (75±13 pg/ml) (Fig. 4).
4. Discussion
Nanoparticles formed from self-assembling peptides, which are close in size to viral particles, are commonly called virus-like particles [19]. These virus-like particles have attracted attention as a convenient approach to vaccine design due to their ability to efficiently deliver epitopes to sensitive cells of the immune system. It was previously shown that recombinant proteins assembled into artificial nanoparticles are highly immunogenic [11, 28]. During nanoparticles assembly process, antigenically active inserted parts are located on the outer surface of the particles, imitating viral particles [19]. And immune response is formed mainly to the inserted components located on the surface of nanoparticles. Several authors have shown that nanoparticles could activate immune system more efficiently than peptides because nanoparticles can be more easily captured and processed by antigen-presenting cells (APCs), thus promoting the maturation of APCs and the immune response activation [29, 30]. It has also been reported that virus-like particles could easily travel to lymph nodes where they presented the antigens to resident APCs and activated T and B cell responses [31].
In the present study, we investigated the efficacy of human DCs activation by artificial nanoparticles carrying CTL epitopes of HCV non-structural proteins. The nanoparticles constructed from mosaic recombinant proteins varied in size from 16 to 80 nm, depending on the number of CTL epitopes localized at the C terminus. The VPL-PE1 protein contained epitopes from NS3 only, while the VPL-PE2 protein, in addition to these epitopes, also contained determinants from NS4a and NS4b. As a result, the VPL-PE2 particle size sharply increased from 25-30 nm to 70-80 nm. This fact indicates the importance of amino acid sequences for the assembly of nanoparticles.
DCs are key antigen-presenting cells in immune response [32-34]. It was shown in model experiments in mice that large nanoparticles (500-2,000 nm) bind well to DCs while smaller nanoparticles (20-200 nm) are more easily transported by DCs to the lymph nodes where they contact with the lymphocytes [35, 36] In our study, nanoparticles with epitopes of HCV proteins varied in size from 25 to 80 nm, which corresponds to smaller nanoparticles (the size of 20-200 nm) that are easily transported to the lymph nodes. In chronic hepatitis C, as shown by a number of authors, DCs largely lose their ability to activate T lymphocytes [37, 38]. Other authors believe that MHC II antigen processing and presentation function is preserved in individuals chronically infected with HCV [39]. It is possible that the problem of weak T cell immune response to candidate vaccine constructs is associated with their ineffective structure.
Currently, biomedicine is actively pursuing research related to the use of virus-like particles as a platform for the construction of recombinant hepatitis C vaccines [40]. Efforts are mainly focused on the use of surface proteins E1 and E2 of HCV. Some positive results have been obtained by a number of authors on chimpanzees [41]. We believe, however, that for more effective vaccine construction, a combination of B and T epitopes of both the structural and non-structural proteins of HCV must be included.
5. Conclusion
In this study, we have created mosaic recombinant proteins capable of self-assembling into nanoparticles and activating DCs that, in turn, successfully stimulated T lymphocytes. Our results indicate that the greatest stimulating effect on T lymphocytes is exerted by DCs activated by 70-80 nm nanoparticles carrying CTL epitopes of HCV NS3, NS4a, and NS4b proteins. Further research in this direction can lead to the development of a promising candidate vaccine against hepatitis C.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the Russian Foundation for Basic Research (projects 20-04-00705).
Authorship Contributions Statement
Victor Kuprianov: project administration, conceptualization, investigation, writing; Lyudmila Nikolaeva: conceptualization, methodology, investigation, writing; Anna Zykova: investigation, methodology; Anna Dedova: investigation; Artemiy Vakhrameev: investigation; Nikolai Ravin: supervision, writing, editing. All authors agree with the text of the article.
References
- The Polaris Observatory HCV Collaborators. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2 (2017): 161-176.
- Bradley H, Hall EW, Rosenthal EM, et al. Hepatitis C virus prevalence in 50 U.S. states and D.C. by sex, birth cohort, and race: 2013-2016. Hepatol Commun 4 (2020): 355-370.
- Bartenschlager R, Baumert TF, Bukh J, et al. Critical Challenges and emerging opportunities in hepatitis C research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res 248 (2018): 53-62.
- Roche B, Coilly A, Duclos-Vallec JC, Sammuel D. The impact of treatment of hepatitis C with DAAs on occurrence of HCC. Liver Int 38 (2018): 139-145.
- Guarino M, Sessa A, Cossiga V, et al. Special interest group on “Hepatocellular carcinoma and new anti-HCV therapies”of the Italian Association for the Study of the Liver. Direct-acting antivirals and hepatocellular carcinoma in chronic hepatitis C: a few lights and many shadows. World J Gastroenterol 24 (2018): 2582-2595.
- Duncan JD, Urbanowicz RA, Tarr AW, Ball JK. Hepatitis C virus vaccine: challenges and prospects. Vaccines 8 (2020): 90.
- Echeverria N, Comas V, Aldunate F, et al. In the era of rapid mRNA-based vaccines: Why is there no effective hepatitis C virus vaccine yet? World J Hepatol 13 (2021): 1234-1268.
- Christiansen D., Earnest-Silveira L., Grubor-Bauk B., et al. Pre-clinical evaluation of a quadrivalent HCV VLP vaccine in pigs following microneedle delivery. Scientific Reports 9 (2019): 9251.
- Guest JD, Pierce BG. Structure-based and rational design of a hepatitis C virus vaccine. Viruses 13 (2021): 837.
- Jahangirian E, Jamal GA, Nouroozi M, Mohammadpour A. A reverse vaccinology and immunoinformatics approach for designing a multiepitope vaccine against SARS-CoV-2. Immunogenetics 5 (2021).
- Babapoor S, Neef T, Mittelholzer C, et al. A novel vaccine using nanoparticle platformto present immunogenic M2e against avian influenza infection. Hindawi Publishing Corporation Influenza Research and Treatment 2011 (2011): 126794.
- Kuprianov VV, Nikolaeva LI, Zykova AA, et al. Combination of three adjuvants enhances the immunogenicity of a recombinant protein containing the CTL epitopes of non-structural proteins of hepatitis C virus. Virus Research 284 (2020): 197984.
- Lauer GM, Barnes E, Lucas M, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterol 127 (2004): 924-936.
- Urbani S, Uggeri J, Matsuura Y, et al. Identification of immunodominant hepatitis C virus (HCV)-specific cytotoxic T-cell epitopes by stimulation with endogenously synthesized HCV antigens. Hepatology 33 (2001): 1533-1543.
- Guo Z, Zhang H, Rao H, et al. DCs pulsed with novel HLA-A2-restricted CTL epitopes against hepatitis C virus induced a broadly reactive anti-HCV-specific T lymphocyte response 7 (2012): e38390.
- Alexander J, Guercio MF, Maewal A, et al. Linear PADRE T-helper epitope and carbohydrate B-cell epitope conjugates induce specific high titer IgG antibody responses. J Immunol 164 (2000): 1625-1633.
- Zhou L-J, Tedder TF. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Natl. Acad. Sci.(US). Immunology 93 (1996): 2588-2592.
- De Filette M., Martens W, Smet A. et al. Universal influenza A M2e-HBc vaccine protects against disease even in the presence of pre-existing anti-HBc antibodies. Vaccine 26 (2008): 6503-6507.
- Raman S, Machaidze G, Lustig A, et al. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomedicine: Nanotechnology, Biology, and Medicine 2 (2006): 95-102.
- Thimme R, Oldach D, Chang K, et al. Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med 194 (2001): 1395-1406.
- Neumann-Haefelin C, Timm J, Spangenberg HC, et al. Virological and immunological determinants of intrahepatic virus-specific CD8+ T-cell failure in chronic hepatitis C virus infection. Hepatology 47 (2008): 1824-1836.
- Ward S, Lauer G, Isba R, et al. Cellular immune responses against hepatitis C virus: the evidence base. Clin Exp Immunol 128 (2002): 195-203.
- Fournillier A, Dupeyrot P, Martin P, et al. Potent induction of T-cell mediated responses following in vitro and in vivo vaccination with hepatitis C virus multiepitope long peptides. Vaccine 24 (2006): 3153-3164.
- Thimme R, Bukh J, Spangenberg HC, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci (US) 99 (2002): 15661-15668.
- Day CL, Lauer GM, Robbins GK, et al. Broad specificity of virus-specific CD4+ T-helper cell responses in resolved hepatitis C virus infection. J Virol 76 (2002): 12584-12595.
- Chang KM, Rehermann B, McHutchison JG, et al. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest 100 (1997): 2376-2385.
- Huang XJ, Lu X, Lei YF, et al. Cellular immunogenicity of a multi-epitope peptide vaccine candidate based on hepatitis C virus NS5A, NS4B and core proteins in HHD-2 mice. J Virol Methods 189 (2013): 47-52.
- Kaba SA, Brando C, Guo Q, et al. A non-adjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J Immunol 183 (2009): 7268-7277.
- Babensee JE. Interaction of dendritic cells with biomaterials. Semin Immunol 20 (2008): 101-108.
- Reddy ST, Swartz MA, Hubbell JA. Targeting dendritic cells with biomaterials: developing the next generation of vaccines. Trends Immunol 27 (2006): 573-379.
- Cubas R, Zhang S, Kwon S, et al. Virus-like particle (VLP) lymphatic trafficking and immune response generation after immunization by different routes. J Immunother 32 (2009): 118-128.
- Hart DNJ. Dendritic cells: uique leukocyte populations which control the primary immune response. Blood 90 (1997): 3245-3287.
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature 392 (1998): 245-252.
- Gluckman J-C, Canque B, Rosenzwajg M. Dendritic cells: a complex simplicity. Transpantation 73 (2002): S3-S6.
- Manolova V, Flace A, Bauer M, et al. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol 38 (2008): 1404-1413.
- Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol 10 (2010): 787-796.
- Tsubouchi E, Akbar S, Horiike N, Onji M. Infection and dysfunction of circulating blood dendritic cells and their subsets in chronic hepatitis C virus infection. J Gastroenterol 39 (2004): 754-762.
- MacDonald AJ, Semper AE, Libri NA, Rosenberg WMC. Monocyte-derived dendritic cell function in chronic hepatitis C is impaired at physiological numbers of dendritic cells. Clin Exp Immunol 148 (2007): 494-500.
- Canaday DH, Burant CJ, Jones L, et al. Preserved MHC-II antigen processing and presentation function in chronic HCV infection. Cell Immunol 266 (2011): 187-191.
- Bellier B. and Klatzmann D., Virus-like particle-based vaccines against hepatitis C virus Expert Rev. Vaccines 12 (2013): 143-154.
- Elmowalid G.A., Qiao M., Jeong S.-H., et al. Immunization with hepatitis C virus-like particles results in control of hepatitis C virus infection in chimpanzees. Proc. Natl Acad. Sci. USA. 104 (2007): 8427-8432.