The Detection of Bornavirus in Birds, Mammals and Snakes by Sandwich ELISA of Bornaviral Matrix Protein
Article Information
Josephine Marie McHugh, Siwo Rel de Kloet*
Animal Genetics Inc., 3382 Capital Circle NE, Tallahassee FL 32308, USA
*Corresponding Author: Dr. Siwo R. de Kloet, Animal Genetics Inc., 3382 Capital Circle NE, Tallahassee FL 32308 USA
Received: 21 September 2021; Accepted: 27 September 2021; Published: 07 October 2021
Citation: Josephine Marie McHugh, Siwo Rel de Kloet. The Detection of Bornavirus in Birds, Mammals and Snakes by Sandwich ELISA of Bornaviral Matrix Protein. Archives of Veterinary Science and Medicine 4 (2021): 85-102.
Share at FacebookAbstract
This study describes a further analysis of the detection of bornaviral antigens by sandwich ELISA. The results show that using chicken anti-psittacine bornaviral P16 matrix protein antibody for capture and rabbit antibody to this protein for detection, recombinant matrix protein of mammalian as well as of distantly related snake bornavirus can be detected, suggesting that a single procedure can be used for the diagnosis of highly divergent bornavirus infections.
Keywords
Sandwich ELISA; Orthobornavirus; Matrix protein
Sandwich ELISA articles; Orthobornavirus articles; Matrix protein articles
Article Details
Abbreviations:
ABG = avian bornaviral ganglioneuritis; ABV = avian bornavirus; AFG = African grey parrot parrot (Psittacus erithacus); AQABV = aquatic bird bornavirus; BSA = bovine serum albumen; CBV = canary (Serinus canaria) bornavirus; EDTA = ethylene diamino tetraacetic acid; Elisa = enzyme-linked immunosorbent assay; ESBV estrildid (Estrilda troglodytes) bornavirus;GLY = glycoprotein; HRP = horse radish peroxidase; LGSBV = Loveridges gartersnake (Elapsoidea loveridgei) bornavirus; MBP = maltose binding protein; MBV = mammalian bornavirus; MWD = macaw wasting disease; PBS = 0.15 M NaCl in 0.02 M sodium phosphate buffer pH 7.4; PDD = proventricular dilatation disease; PMSF = phenylmethylsulfonyl fluoride; POL = bornavirus RNA polymerase; PsBV = Psittaciform bornavirus; PVDF = polyvinylidene difluoride; SC = sun conure (Aratinga solstitialis); TBST = 0.15 M NaCl containing 0.02 M tris-HCl buffer pH 7.0 and 0.05% Tween 20; TBSTM = TBST containing 5% fat free dried milk; TMB = 3,3′,5,5′-tetramethylbenzidine; VSBV = variegated squirrel (Sciurus variegatoides) bornavirus;VSV = vesicular stomatitis virus
1. Introduction
Psittaciform orthobornavirus (PsBV) (order Mononegavirales, family Bornaviridae, genus Orthobornavirus, [1-3]) is considered to be the cause of an often fatal disease of parrots and parakeets [4, 5], originally known as macaw wasting disease (MWD), but also known as proventricular dilatation disease (PDD) or avian bornaviral ganglioneuritis (ABG) [6]. The disease is mostly recognized by neurological symptoms [7, 8], such as unstable gait and feather picking but often also by intestinal symptoms such as inflammation of the proventriculus [7] causing intestinal blockage, leading to starvation. The disease is therefore often first recognized by the occurrence of undigested seed in fecal matter. Many birds, infected with bornavirus, are however symptom free and can live a long, healthy life [9, 10].Bornavirus is a negative-stranded RNA virus, with a genome of approximately 8900 nts [11, 12]. Different strains occur in birds, such as waterfowl [13], songbirds and [14] psittaciformes [4, 5], mammals such as horse and sheep [15, 16] as well as snakes [17, 18]. Related viruses are found in fungi [19], pythons [20] and fish [21]. Currently two species of psittacine bornavirus are recognized containing a total of eight strains [1-3]. The transcriptome consists of three transcriptional units [11, 12], carrying six genes coding for 1) P40, a nucleoprotein, 2) P10, a cofactor for transcription, 3) P24, a phosphoprotein, 4) P16, a matrix protein, 5) P57, a glycosylated capsid protein and 6) a RNA dependent RNA polymerase. Gene expression involves truncation of some gene transcripts (nucleoprotein, phosphoprotein) as well as splicing of the primary transcripts (e.g. nucleoprotein, glycoprotein and polymerase), using the host gene splicing apparatus [11, 12]. Gene expression of the proteins shows a gradient in which the most 3’gene (the nucleoprotein) is expressed at a higher copy number than the 5’-gene for the polymerase. Copies of segments of bornavirus occur frequently as endogenous viral elements in the genome of mammals and snakes [22, 23] but are scarce in birds [24]. Virus-like structures have been observed in birds infected with bornavirus [25] and psittaciform bornavirus can be cultured from infected tissue segments in Japanese quail cells [26] or duck embryo cells [27], but generally not in mammalian cells [28]. However, the complete physical purification of psittaciform bornavirus has not been described and although the purification of mammalian bornavirus by CsCl density gradient centrifugation [29], affinity chromatography with “virobeads” [30] and tandem affinity purification [31] has been reported, the actual quantitative molecular composition of the particles of this virus has however also not been described, except for the observation that the molecular ratio between the P24 phosphoprotein and the P40 nucleoprotein was much higher in persistently infected tissues than in acutely infected cells [32, 33].Bornavirus infection in psittaciformes, an avian family which contains many endangered species, is primarily a disease of captive birds and has not been found with certainty in truly wild or feral populations [34-36]. Although, the occurrence of psittaciform specific strains suggests strongly that such strains exist naturally, under natural conditions the virus may have little chance of inducing disease. The bornaviruses occurring in waterfowl and snakes are often found in animals suffering from neurological problems. The chance that psittaciform borna disease may affect species such as the Spix macaw (Cyanopsitta spixii) which currently only survives in captivity [e.g. 37] or the Kakapo (Strigops habloptilus) which has a very limited geographical distribution on a few islands in New Zealand [38] makes that bornavirus can be an important factor in the survival of such species, especially those which are only maintained in captivity or semicaptivity. A major problem is that the mode by which birds in captivity transmit the infection is not known, but phylogenetic studies have suggested that horizontal transmission must occur [39]. In most cases a virus is transmitted between hosts through involvement of cellular receptors which specifically bind a component of the viral structure. However, in bornavirus, such cellular receptors have not been described and host-specificity is at least in part the result of the specific interaction between the nuclear localization factor of the nucleoprotein and elements in the nuclei of the cells of a receptive, avian or mammalian host [28]. “Contact” and oral-fecal transmission have been suspected for a long time as the cause of horizontal spread of the infection, but could not be proven [40]. Transmission through skin damage is suspected [41], which makes that in addition to wounds, also behavioural issues such as “mutual preening” of the plumage can be involved, especially since feather calami can be a rich source of bornavirus [42]. Nevertheless, infected and uninfected birds can be housed together for years without the latter becoming infected. Experimental infections are usually done by injection of infected tissue homogenates or cell cultures [43, 44], but the virus can also be vertically transmitted through eggs [45, 46] and suspected cases of “spontaneous infection” may well be the result of birds having asymptomatic parents [47] or that the diagnostic procedure was not sensitive enough to detect the infection in these parents. Finally, our knowledge with regard to bornavirus is mostly limited to viruses occurring in mammals and birds. Almost nothing is known about bornaviruses in fish, amphibians, reptiles and invertebrates and their role in borna disease.Bornavirus infections used to be diagnosed mainly by their clinical signs or by virus culture, but the difficulties in the latter procedure as well as the occurrence of many asymptomatic carriers of the virus have made molecular diagnosis by reverse transcriptase PCR of the viral RNA and serological analysis by ELISA for the detection of specific serum antibodies the current methods of choice. For ante mortem RNA analysis, throat, cloacal swabs or extracts of feather calami can be used as a relatively noninvasive source of the RNA [42]. The use of antibodies as diagnostic material is hampered by the fact that some of the infected birds fail to form antibodies to bornaviral antigens [48]. In addition blood collection under natural field conditions can interfere with the subsequent analysis, due to poor stability of the samples under such conditions, often leading to false positives [49] or negatives. Diagnosis through RT-PCR analysis suffers from the instability of RNA, which especially in field work may present difficulties, due to the time and conditions involved between collection of the diagnostic material and the analysis. PCR can be influenced by mutations anywhere in the amplicon, giving rise to primer-mismatch or secondary structure alterations which give rise to false negatives [50]. In addition, reverse transcriptase has been found to suffer from false priming [51, 52]. Finally, the multitude of strains makes it virtually impossible to design a single primer set to detect every strain of bornavirus.Viral proteins are direct phenotypic determinants of viral structure and biochemical activity.Therefore the analysis of viral protein is more indicative of active viral presence than nucleic acids, which can fail to express their genetic content. In addition, viral protein antigens are generally more stable than nucleic acids [53] and could therefore be more useful for analytical studies, especially in cases where the logistics form a difficulty in the useful preservation of RNA in biological samples, such as collecting diagnostic material in the native habitat of animals being studied, which may be in the case of parrots often tropical, adverse climates.Recently we have described the detection of psittacine bornaviral antigens in feather calami and cloacal swabs through analysis by sandwich ELISA, using chicken polyclonal antibodies as capture antibodies and rabbit polyclonal antibodies for the detection of the P24 phosphoprotein and P16 matrix protein antigens [54]. In the present study we describe the molecular procedures used in the diagnosis of a bornavirus infection by the sandwich ELISA procedure more in detail and report that the matrix protein can be used as a sensitive diagnostic marker for detecting infection of a wide variety of animals with bornavirus.
2. Materials and Methods
2.1 Bird maintenance, determination of infection with avian bornavirus and collection of tissues and bloodMost birds included in these studies were maintained in the private collection of the senior author (S.R.de K.) as described in previous publications [42]. Additional bird tissues, feathers and blood were obtained from other, local, breeders. Tissues were obtained from birds which had died from PDD or from other causes, such as fighting, predation, accidents, etc. No birds were euthanized for this investigation. Feathers (contour and wing cover) were obtained by plucking from dead, as well as living birds. Cloacal swabs were obtained using cotton swabs. No anesthesia was used. Infection with bornavirus was detected by RT-PCR of feather calamus RNA as described in an earlier publication [42] using two primer sets that amplify a) the P10 gene and b) a segment of the polymerase gene. Blood was collected from the brachial vein using 20 ml EDTA coated glass capillaries (Cat # 19.447, Sarstedt, Newton, NC, USA). For routine Elisa [42] the contents of the capillaries were immediately diluted 10 fold in 180 ml of 0.15 M NaCl containing 0.025 M Na2-EDTA pH 8.0 (buffer A) and centrifuged for 10 min in an Eppendorf minifuge at 2000g at 4oC to obtain plasma.
2.2 Synthesis of recombinant bornavirus proteins for use in sandwich ELISA
Recombinant bornavirus proteins, used in this study were synthesized in E. coli as conjugates with maltose binding protein (MBP), as described earlier [42]. The ABV5-P16 and ABV5-P24 antigens were synthesized for us by Genscript (Piscataway, NJ, USA). The amino acid sequences of the proteins used in this study were obtained from the Genbank database. The nucleotide sequences used for expressing the recombinant proteins were either the natural sequences as given in the database or “codon optimized” for us by IDT (Coralville, IA, USA). The proteins and their Genbank accession numbers are given in Table 1.|
Sno. |
|
|
Genbank protein acc. nr. |
|
1 |
ABV2 |
P24 phosphoprotein |
ADY39269. |
|
2 |
ABV2 | P16 matrix protein | ACS32308.1 |
|
3 |
ABV5 |
P24 phosphoprotein |
QAT79256 |
|
4 |
ABV5 |
P16 matrix protein |
QAT79257 |
|
5 |
AQABV1 |
P24 phosphoprotein |
KU748788 |
|
6 |
AQABV1 |
P16 matrix protein |
MK966418 |
|
7 |
MBV1 |
P24 phosphoprotein |
QBY26072 |
|
8 |
MBV1 |
P16 matrix protein |
QBY26070 |
|
9 |
LGSBV1 |
P24 phosphoprotein |
YP_009055060 |
|
10 |
LGSBV1 |
P16 matrix protein |
YP_009055061 |
Table 1: Genbank Accession nrs of recombinant bornavirus proteins generated in the present investigation. ABV = avian (psittacine) bornavirus; AQABV = aquatic bird bornavirus; MBV = mammalian bornavirus; LGSBV = Loveridges garter snake.
2.3 Induction in chickens and isolation of polyclonal anti-avian bornavirus antibodies from egg yolk
One year old Bantam chickens were immunized with 100 mg P24 or P16 conjugated to maltose binding protein (MBP), prepared as described before [42] in 100 ml TBS (0.15 M NaCl, containing 0.02 M tris-HCl buffer pH 7.0) and 100 ml complete Freunds adjuvant (Cat# 77140, Thermofisher Scientific) by injection at two different places in the breast muscle. The immunization was repeated after 2 weeks using incomplete Freunds adjuvant (Cat# 77145, Thermofisher Scientific). Eggs were collected beginning at 2 weeks subsequent to the last immunization. Polyclonal antibodies were isolated from chicken eggs by the polyethylene glycol (PEG) procedure [55-57], followed by affinity purification using thiophilic gel chromatography [58]. The concentration of the IgY was measured by the Bradford assay [59], using purified bovine IgG as the standard and the solution diluted in TBST or concentrated by ultrafiltration (Centricon YM 10, Millipore, Burlington, MA, USA) until a final concentration of 2 mg per ml was reached. The titers of the antibody preparations were: anti-P24 = 5,000; anti-P16 = 10,000.
2.4 Generation of rabbit antibodies against recombinant psittacine bornavirus antigens
Antibodies against the P24 and P16 proteins of parrot bornavirus strain 2 (PSBV2) were generated for us in rabbits by Genscript The accession numbers of the bornaviral antigens were as described above. The titers of the antibodies were as given by the manufacturer, 100,000. The antigens were used for immunization after removal of the fusion partner (MBP) and prepared for us by Genscript.
2.5 Sandwich Elisa for the detection of bornaviral antigens
Sandwich ELISA assays [60, 61] for detection of the presence of psittaciform borna virus P24 and P16 antigens in tissue homogenate, cloacal swabs or feather calami using chicken and rabbit polyclonal antibodies were carried out using the protocol provided by Abcam (Cambridge, Mass, USA) with some modifications as described previously [54]. The entire procedure was as follows: wells of an ELISA plate were filled with 50 ml 1,000 fold diluted chicken anti-ABV P24 or P16 IgY, prepared as described above as the capture antibody. Plates were shaken for an hour at room temperature, washed with TBST, blocked for an hour with 100 ml TBSTM and washed again with TBST. Subsequently, recombinant antigens, feather or cloacal swab extracts, prepared as described before [54], dissolved in 50 ml TBSTM were added in the amounts indicated in the figures, and shaking continued for another hour. After washing with TBST, 50 ml of detection rabbit polyclonal anti-P24 or -P16 antibody, diluted 2,000 fold in TBSTM, was added and shaking continued for another hour. The plates were washed again with TBST, incubated for an hour with 50 ml 2,000 fold goat anti-rabbit antibody conjugated to horse radish peroxidase (Cat# A140-110P, Bethyl, Montgomery, TX, USA), diluted in TBSTM, washed again with TBST and finally incubated at room temperature (approx. 22oC) for approximately 15 min with 50 ml of non-precipitating, stabilized TMB solution (Cat# TM1999, Scytek Laboratories, Logan, UT, USA). The reaction was stopped with 50 ml 0.5 N HCl, and the optical density was measured at 450 nm, using a microplate reader (Tecan, Mannedorf, Switzerland). If needed, photographs of the plates were made prior to the addition of the 0.5N HCl. For chemiluminescent detection, SuperSignalR ELISA Pico Chemiluminescent Substrate was used according to the recommendations of the manufacturer (Thermoscientific/Pierce Biotechnology, Rockford IL, USA).
3. Results
3.1 Bornavirus antigen detection by sandwich ELISA
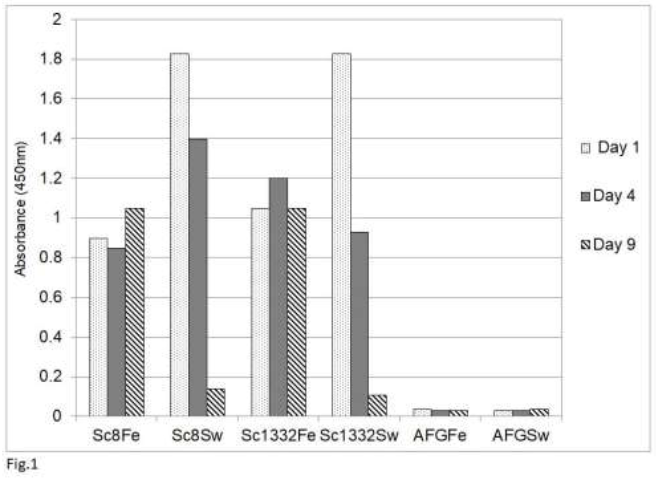
Our previous study [54] showed that an analysis by sandwich ELISA of tissue homogenates of psittaciformes infected with psittacine bornavirus 2 demonstrated the presence of the P24 phosphoprotein and the P16 matrix protein in these homogenates as well as in feather calami and cloacal swabs. The glycoprotein (P27, P29) could not be detected because tissues of persistently infected parrots contain very little of the glycosylated or nonglycosylated forms of this protein [42, 48]. The results of this study also showed that the rabbit anti-P10 antibody cross-reacts with a number host proteins and could therefore not be used. Finally, the RNA polymerase was not used because of its low level of expression in bornavirus [11]. Using the rabbit antibody against the C-terminal 330 amino acid long fragment of the P40 nucleoprotein, the detection of the P40 nucleoprotein was also rather poor, although Western blotting analysis showed the abundant presence of P40 in infected tissues [42]. A poor detection by sandwich ELISA analysis may be possible if the chicken and rabbit antibodies recognize the same epitopes on the P40 nucleoprotein (Abcam, Genscript pers. comm.). This would mean that P40 proteins which are bound to the chicken capture antibodies on the plate, cannot bind to the rabbit antibodies used as the detection antibodies. The P24 phosphoprotein and the P16 matrix protein are therefore the preferred antigens for bornaviral detection by the sandwich ELISA procedure. An investigation of the stability of P24 and P16 in cloacal swabs, fecal matter or feather calami of several parrots (Figure 1) showed that both antigens (shown for P16 only) become undetectable in cloacal swabs or
Figure 1: Stability of avian bornavirus proteins in fecal matter or feather calami of two sun conures (Sc8 and Sc 1332) upon storage at room temperature as measured by the sandwich ELISA of bornavirus P16 matrix protein. Feather (Fe) and fecal swabs (Sw) were collected and stored at ambient temperature (approx. 25oC) for the indicated number of days. AFG is material from an uninfected African grey parrot (Psittacus erithacus).
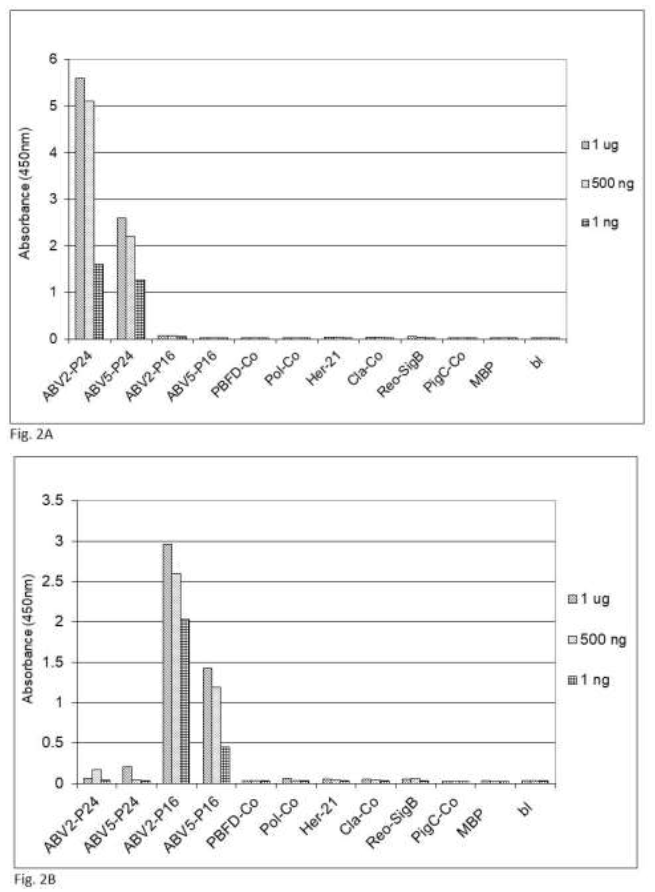
Figure 2: Specifity of the sandwich ELISA for psittacine bornavirus. Sandwich ELISA using chicken capture antibodies and rabbit detection antibodies against (2A) bornavirus ABV2 P24 phosphoprotein or (2B) ABV2 P16 matrix protein were used to detect reactivity against recombinant forms of protein component from frequently occurring parrot pathogens. PBFD-Co = coat protein of psittacine beak and feather virus (AAS93283); Pol-Co = coat protein of avian polyomavirus (AF241168); Her-21 = glycoprotein E of psittacine alpha herpesvirus1 (NP_944450); Cla-co = outer membrane protein B of Chlamydia psittacae (EPJ24186); Reo-SigB = sigma B protein of psittacine reovirus (ACD43460); PigC-Co = coat protein of pigeon circovirus (ACJ14531).
fecal matter after a week of storage at ambient temperature, which means that feather calami are the most practical noninvasive source material for the identification of psittacine bornavirus antigenic material under difficult environmental circumstances. Further investigations showed that the described sandwich ELISA procedure is specific for bornavirus since uninfected birds or birds infected with other frequently occurring diseases such as PBFD, avian polyomavirus, avian herpes virus, psittacine reovirus or Chlamydia psittaci or isolated recombinant proteins [described in earlier papers, 62, 63] from these organisms at a concentration of 1 ng to 1 mg did not give a signal (Figure 2).
The maximum sensitivity of the detection of the P16 matrix protein, using the TMB color reaction is about 1 ng, using chemiluminescent detection the sensitivity could be improved to approximately 100 pg (data not shown). Although most strains of parrot bornavirus belong to one species (parrot bornavirus 1) recently another strain [parrot bornavirus 5 (PaBV5)] has been detected [64-67], of which the nucleotide sequence more closely resembles the sequence of other avian non-parrot and mammalian bornaviruses (Table 2, Figure 3) and which phylogenetically groups (Figure 3) with these other avian bornaviruses and is therefore considered to belong to another bornavirus species: psittacine bornavirus 2.
|
|
ABV4 |
ABV5 |
AQABV1 |
MBV1 |
LGSBV1 |
|
P16 |
98% |
87% |
87% |
83% |
71% |
|
POLFR |
94% |
77% |
80% |
81% |
71% |
|
P40 |
95% |
70% |
77% |
75% |
68% |
|
POL |
93% |
68% |
70% |
68% |
58% |
|
GLY |
89% |
67% |
72% |
65% |
62% |
|
P24 |
95% |
70% |
69% |
62% |
45% |
|
P10 |
76% |
52% |
53% |
45% |
34% |
Table 2: Sequence conservation of bornavirus proteins of different bornaviruses. Values are as compared to the sequence of ABV2, measured as percentage of amino acids in identical positions. ABV = psittacine bornavirus; AQABV = aquatic bird bornavirus; MBV = mammalian bornavirus; LGSBV = Loveridges garter snake bornavirus. POLFR is a highly conserved fragment (AA543 -716) of bornaviral RNA polymerase.
Figure 3: Neighbour-joining tree of the amino acid sequences of the P16 matrix proteins of different bornaviral strains. Sequences were aligned in Clustal X and a neighbour-joining tree was generated in MEGA 5.2. ABV = psittacine (avian) bornavirus (ABV1 = AFN70791; ABV2 = ACS32308.1; ABV3 = ACH61939; ABV4 = ADY39270; ABV5 = BAV57485; ABV7 = YP_009268902); AQABV = aquatic bird bornavirus (AQABV1 = QGZ07386; AQABV2 = YP_009268914); CBV = canary bornavirus (CBV1 = YP_009268908; CBV2 = YP_009165495; CBV3 = YP_009041459); ESBV = estrilde bornavirus (ESBV1 = YP_009505426); LGSBV = Loveridges garter snake bornavirus (LGSBV1 = YP_009055061); MBV = mammalian bornavirus (MBV1 = NP_042022; MBV2 = YP_009268920); VSBV = variegated squirrel bornavirus (VSBV1 = YP_009269416).
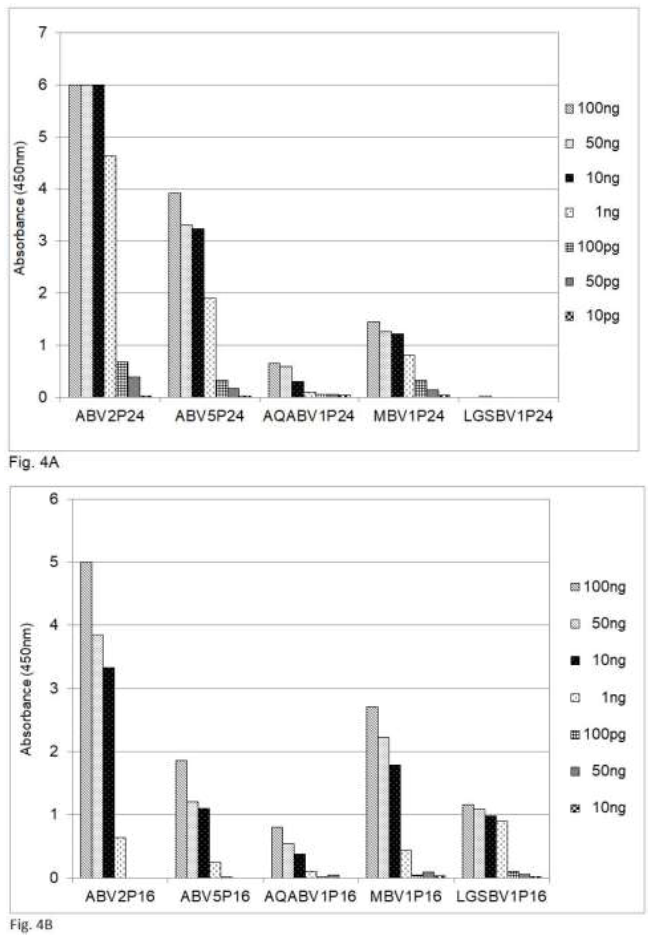
A comparison of the amino acid sequences shows that the sequences of the P24 phosphoprotein and the P16 matrix protein of PaBV5 and PaBV2, which belongs to psittacine bornavirus 1, are approximately 70 and 87 per cent identical, respectively (Table 2). Recently bornaviruses or borna-like viruses have also been discovered in snakes and fish. The viruses described by Stenglein et al. [17] in the Loveridges garter snake (Elapsoidea loveridgei) and by Pfaff and Rubbenstroth [18] in Caribbean watersnakes (Tretanorhinus variabilis) and Mexican rattle snakes (Crotalus molossus nigrescens) have a gene order similar to that of the other, avian or mammalian, bornaviruses, whereas the viruses in certain pythons are different, at least in this respect [20]. The data in Table 2 show that the amino acid sequence of the P16 matrix protein is up to 71% conserved between the Loveridges garter snake bornavirus and the P16 protein of the parrots and mammals, whereas the amino acid sequence of the P24 is much less conserved (45%). The data in Figure 4 show that in an analysis by sandwich ELISA the mammalian.
Figure 4: Detection of bornavirus antigens from birds, mammals and a snake by sandwich ELISA, using capture antibodies from chickens and detection antibodies from rabbits against the P24 phosphoprotein and the P16 matrix protein. ABV = psittacine bornavirus; AQABV = aquatic bird bornavirus; MBV = mammalian bornavirus; LGSBV = Loveridges garter snake bornavirus. 4A = P24 phosphoprotein; 4B = P16 matrix protein.
Bornavirus (MBV), the bornavirus found in aquatic birds (AQABV) and the parrot bornavirus ABV5 as well as the bornavirus of the Loveridges garter snake (LGSBV) can be detected by using capture and detection antibodies against the ABV2 P16 matrix protein (Figure 4B) from chicken resp. rabbits, whereas the antibodies against the ABV2 P24 phosphoprotein (Figure 4A) fail to detect the snake bornavirus.
4. Discussion
Bornavirus has long been suspected of being a pathogenic virus in humans. Although the early data were controversial, recently the infection of caretakers with bornavirus from captive squirrels, has demonstrated that such infections are real and can be fatal [68]. The present study was mainly undertaken in order to improve methods to collect diagnostic samples from Psittaciformes that result in accurate data with regard to infection with avian bornavirus, which are not dependent on the use of RT-PCR, because of the instability of RNA due to logistical circumstances, which may occur during collection and transportation of diagnostic samples. In addition it is difficult, if not impossible to design RT-PCR primers which cover all different bornaviruses [69]. Therefore, these conditions call therefore for the development of other diagnostic procedures for the detection of a bornavirus infection, which are less sequence dependent. The results obtained in this study show that the colorimetric or chemiluminescent detection of P16 matrix protein by sandwich ELISA using chicken capture and rabbit detection antibodies against the P16 matrix protein of ABV2 in parrots, satisfies this requirement and can be extended to detecting bornavirus in mammals and snakes, thereby providing for one single diagnostic procedure to detect bornavirus in a wide variety of hosts. An additional advantage of using the P16 matrix protein antigen is that thus far, in uninfected birds, we have not detected any antigen that may cross react with chicken or rabbit antibodies against the P16 matrix protein of bornavirus and that the detection is specific for bornavirus, for no signal is obtained with recombinant antigen for some of the common pathogens. It is however unknown whether the same is the case for snakes and other animals. A similar procedure using the P24 phosphoprotein can detect ABV5 and mammalian bornavirus, but fails to detect the snake bornavirus. As described above, the other antigens, the P40 nucleoprotein, the P57 glycoprotein, the P10 X protein or the L(RNA polymerase) cannot be used for various reasons such as limited presence or cross reactivity with other non-bornaviral proteins. The amino acid sequences of the P24 phosphoprotein and the P16 matrix protein of the bornaviruses infecting parrots and the snake are resp. 45% and 70% identical. Earlier studies by Wilson and Prager [70, 71] and others have shown that antibodies can generally cross-react with homologous antigens which have less than approximately 35% sequence dissimilarity. The results of the present investigation agree also with those of Zimmermann et al, [69] that antibodies against bornaviral antigens of one species (e.g. found in birds) may show extensive cross reactivity with homologous antigens from a wide variety of other strains. Apparently this observation also holds true when two antibodies are involved, as in the sandwich ELISA procedure.
Recently viruses which are related to bornaviruses have been detected in pythons [20], in a fish [21] and possibly fungi [19]. A major difference between these latter viruses and the orthobornavirus found in birds, mammals and the Loveridges garter snake is that the location of the sequences of the glycoprotein gene and the matrix protein have been interchanged. These viruses are therefore considered to belong to other genuses: Carbovirus, resp Cultervirus [3]. The P16 matrix protein amino acid sequences of the Carbo- and Culterviruses show however a rather limited sequence similarity (less than 25%) with the sequences of the true bornaviruses, which makes it unlikely that the proposed sandwich ELISA could be used for the detection of Carbo- or Culterviruses.
Using non-precipitating, stabilized TMB solution (Cat# TM1999, Scytek Laboratories, Logan, UT, USA) for the colorimetric detection, the maximal sensitivity of this procedure was in the 1 ng range. Using chemiluminescent detection, the sensitivity could be improved to100 pg. The usefulness of the sandwich ELISA procedure for the detection of the P24 phosphoprotein and P16 matrix protein in fecal matter or cloacal swabs is somewhat diminished due to the reduced survival of these proteins in the materials and calls for procedures which improve their stability in such materials. In birds, this leaves feather calami as the most useful, relatively noninvasive, diagnostic source, a conclusion reached earlier by others in studies on waterfowl and influenza virus [72]. Whereas, in other animals the usefulness of preservatives in fecal, cloacal swabs or coanal samples will have to be studied.
5. Conclusion
The present investigation describes a sandwich ELISA procedure that uses chicken antibodies against the psittacine bornavirus matrix protein for capture and rabbit antibodies against antigen for detection that can detect the bornavirus matrix protein of many strains of the virus, opening the way to the development of a single procedure for the specific detection of avian, mammalian and snake bornavirus.
Conflict of Interest
The authors declare no conflict of interest.
References
- Kuhn JH, Dürrwald R, Bào Y, et al. Taxonomic reorganization of the family Bornaviridae. ArchVirol 160 (2015): 621-632.
- Amarasinghe GK,Aréchiga Ceballos NG,BanyardAC,et al. Taxonomy of the order Mononegavirales: update 2018. Arch Virol 163 (2018): 2283-2294.
- Rubbenstroth D,Briese T,Dürrwald R, etal. ICTV virus taxonomy profile: BORNAVIRIDAE. J Gen Virol 102 (2021): 001613 D
- HonkavuoriKS, Shivaprasad HL, Williams BL, et al. Novel bornavirus in psittacine birds with proventricular dilatation disease. Emerg Infect Dis 14 (2008): 1883-1886.
- KistlerAL,Gancz A, Clubb S, et al. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol J 5 (2008): 88.
- Gregory C. A review of proventricular dilatation syndrome.J Assoc Avian Vet 8 (1994): 69-75.
- Hoppes SM, Shivaprasad HL, et al. Proventricular dilatation disease; Diagnosis, Pathology, Prevalence, and control. The Veterinary Clinics of North America Exotic Animal Practice 16 (2013): 339-355.
- Fluck A,Enderlein D, Piepenbring A, et al. Correlation ofavianbornavirus-specific anti-bodies and viral ribonucleic acid shedding with neurologicalsignsand feather damage-ing behaviour in psittacinebirds. Vet Rec 184 (2019): 476.
- De KloetSR,DorresteinGM. Presence of avian bornavirus RNA and anti-avian bornavirus antibodies in apparently healthy macaws. Avian Dis53 (2009): 568-573.
- Tizard I, Shivaprasad HL, Guo J, et al. The pathogenesis of proventricular dilatation disease. Anim Health Res Rev 17 (2016): 110-126.
- Briese T, Schneemann A, Lewis AJ, et al. Genomic organization of Borna disease virus. Proc Natl Acad Sci USA 91 (1994): 4362-4366.
- de la Torre JC. Molecular biology of Borna disease virus and persistence. Front Biosci 7 (2002): 569-579.
- Delnatte P, Ojkic D, Delay J, et al. Pathology and diagnosis of avianbornavirusinfection in wild Canada geese (Branta canadensis), trumpeter swans (Cygnus buccinator) and mute swans (Cygnus olor) in Canada: a retrospective study. Avian Pathol 42 (2013): 114-128.
- Rubbenstroth D, Rinder M, Stein M, et al. Avian bornaviruses are widely distributed in canary birds (Serinus canaria f. domestica). Vet Microbiol 165 (2013): 287-295.
- Ludwig H, Bode L, Gosztonyi G. Borna disease: a persistent virus infection of the central nervous system. Prog Med Virol 35 (1988): 107-151.
- Tizard I, Ball J, Stoica G, et al. The pathogenesis of bornaviral diseases inmammals. Anim Health Res Rev 17 (2016): 92-109.
- Stenglein MD,Leavitt EB,Abramovitch MA, et al. Genome Sequence of a Bornavirus Recovered from a Loveridge Garter Snake (Elapsoidea loveridgei).Genome Announc 2 (2014): e00779-e14.
- Pfaff F, Rubbenstroth D. Two novel bornaviruses identified in colubrid and viperidsnakes. Arch Virol 166 (2021): 2611-2614.
- Liu L, Xie J, Cheng J, et al. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A 111 (2014): 12205-12210.
- Hyndman TH,Shilton CM,Stenglein MD, et al. Divergent bornaviruses from Australian carpet pythons with neurological disease date the origin of extant Bornaviridae prior to the end-Cretaceous extinction. PLoS Pathog 14 (2018): e1006881.
- Shi M, Lin X-D, Chen X, et al. The evolutionary history of vertebrate RNA viruses. Nature 556 (2018): 197-202.
- Horie M, Tomonaga K. Paleovirology of bornaviruses: What can be learned from molecular fossils of bornaviruses. Virus Res 262 (2019): 2-9.
- Gilbert C, Meik JM, Dashevsky D, et al. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci. B 22 (2014) 281 (1791): 20141122.
- Cui J, Zhao W,Huang Z, et al. Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biology 15 (2014): 539-552.
- Gough RE, Drury SE, Harcourt-Brown NH, et al. Virus-like particles associated with macaw wasting disease.Vet Rec139 (1996): 24.
- Rubbenstroth D, Rinder M, Kaspers B, et al. Efficient isolation of avian bornaviruses (ABV) from naturally infected psittacine birds and identification of a new ABV genotype from a salmon-crested cockatoo (Cacatua moluccensis). Vet Microbiol 161 (2012): 36-42.
- Guo J, Payne S, Zhang S, et al. Avian bornaviruses: diagnosis, isolation, and genotyping. Curr Protoc Microbiol 34 (2014): 15I.1.1-33.
- Komorizono R, Sassa Y, Horie M, et al. Evolutionary Selection of theNuclearLocalizationSignal in the ViralNucleoproteinLeads to Host Adaptation of the Genus Orthobornavirus. Virusses 12 (2020): 1291.
- Zimmermann W, Breter H, Rudolph M, et al. Borna disease virus: immunoelectron microscopic characterization of cell-free virus and further information about the genome. J Virol 68 (1994): 6755-6758.
- SakudoA, Tanaka Y, Ikuta K, et al. Capture of infectious borna disease virus using anionic polymer-coated magnetic beads. Neurosci Lett 494 (2011): 237-239.
- Mayer D,Baginsky S,Schwemmle M. Isolation of viral ribonucleoprotein complexes from infected cells by tandem affinity purification. Proteomics 5 (2005): 4483-4487.
- Watanabe M,Zhong Q,Kobayashi T, et al. Molecular ratio between borna disease viral-p40 and -p24 proteins in infected cells determined by quantitative antigen capture ELISA. Microbiol Immunol 44 (2000): 765-772.
- Schneider U, Naegele M, Staeheli, et al. Active borna disease virus polymerase complex requires a distinct nucleoprotein-to-phosphoprotein ratio but no viral X protein. J Virol 77 (2003): 11781-11789.
- Kessler S,Heenemann K,Krause T,et al. Monitoring of free-ranging and captive Psittaculapopulations in Western Europe for avian bornaviruses, circoviruses and polyomaviruses Avian Pathol 49 (2020): 119-130.
- Encinas-Nagel N,Enderlein D,Piepenbring A, et al. Avian bornavirus in free ranging psittacine birds in Brazil. Emerg Infect Dis 20 (2014): 2103-2106.
- Sassa Y, Bui VN, Saitoh K, et al. Parrot bornavirus-2 and -4 RNA detected inwildbird samples in Japan are phylogenetically adjacent to those found in petbirdsin Japan. Virus Genes 51 (2015): 234-243.
- Fischer D, Neumann D, Purchase C, et al. The use of semen evaluation and assisted reproduction inSpix'smacawsin terms of species conservation.Zoo Biol 33 (2014): 234-244.
- Clout MN.A celebration of kakapo: progress in the conservation of an enigmatic parrot.Notornis 53 (2006): 1-2.
- Rubbenstroth D, Schmidt V, Rinder M, et al. Phylogenetic Analysis Supports Horizontal Transmission as a Driving Force of the Spread of Avian Bornaviruses. PLoS One 11 (2016): e0160936.
- Heffels-Redmann U, Enderlein D, et al. Follow-up investigations on different courses of natural avianbornavirusinfections in psittacines. Avian Dis 56 (2012): 153-159.
- Heckmann J, Enderlein D, Gartner AM, et al. Wounds as the Portal of Entrance for Entrance of Parrot Bornavirus 4 (PaBV-4) and Retrograde Axonal Transport in Experimentally Infected Cockatiels (Nymphicus hollandicus). Avian Diseases 84 (2020): 247-255.
- de Kloet AH, Kerski A, de Kloet SR. Diagnosis of avian Bornavirus infection in psittaciformes by serum antibody detection and reverse transcription assay using feather calami. J Vet Diagn Invest 23 (2011): 421-429.
- Olbert M, Römer-Oberdörfer A, Herden C, et al. Viral vector vaccines expressing nucleoprotein and phosphoprotein genes. Sci Rep 6 (2016): 36840.
- Gray P, Hoppes S, Suchodolski P, et al.Use ofavianbornavirusisolates to induce proventricular dilatation disease in conures. Emerg Infect Dis 16 (2010): 473-479.
- Lierz M, Piepenbring A, Herden C, et al. Verticaltransmissionofavianbornavirusin psittacines. Emerg Infect Dis 17 (2011): 2390-2391.
- Kerski A, de Kloet AH, de Kloet SR. Verticaltransmissionofavianbornavirusin Psittaciformes:avianbornavirusRNA and anti-avianbornavirusantibodies in eggs, embryos, and hatchlings obtained from infected sun conures (Aratinga solstitialis). Avian Dis 56 (2012): 471-478.
- Ogawa H, Sanada Y, Sanada N, et al. Proventricular dilatation disease associated withavianbornavirusinfection in a Citron-crested Cockatoo that was born and hand-reared inJapan. Vet Med Sci 73 (2011): 837-840.
- McHugh JM, de Kloet SR. Discrepancy in the diagnosis of avianbornavirus infection of psittaciformes by protein analysis of feathers calami and enzyme-limked immunosorbent assay of serum antibodies. J Vet Diagn Invest 27 (2015): 150-158.
- Hoye BJ. Variation in postsampling treatment of avian blood affects ecophysiological interpretations. Methods in Ecology and Evolution 3 (2012): 162-167.
- Liu Q, Thorland EC,Sommer Inhibition of PCR amplification by a point mutation downstream of a primer. Biotechniques 22 (1997): 292-294.
- Moison C, Arimondo PB,Guieysse-Peugeot A-L. Commercial reverse transcriptase as source of false-positive strand-specific RNA detection in human cells. Biochimie 93 (2011): 1731-1737.
- Tuiskunen A, Leparc-Goffart I, Boubis L, et al. Self-primingofreversetranscriptaseimpairs strand-specific detection of dengue virusRNA. Gen Virol 91 (2010): 1019-1027.
- Service RF. Researchers close in on ancientdinosaurproteins. Science 355 (2017): 441-442.
- McHugh JM, de Kloet SR Bornavirus antigens in psittaciformes infected with psittaciform bornavirus and their use in diagnostic procedures. Arch Vet Sci Med 3 (2020): 63-90.
- Carlander D,Stålberg J,Larsson Chicken Antibodies: A Clinical Chemistry Perspective. Ups J Med Sci 104 (1999): 179-189.
- Gassmann M, Thömmes P, Weiser T, et al. FASEB J 4 (1990): 2528-2532.
- Polson A, von Wechmar MB, van Regenmortel MH. Isolation of viral IgY antibodies from yolks of immunized hens. Immunol Commun 9 (1980): 475-493.
- Porath J, Maisano F, Belew M. Thiophilic adsorption – a new method for protein fractionation. FEBS Lett 185 (1985): 306-310. >
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 (1976): 248-254.
- Crowther JR. The ELISA Guidebook. Springer Protocols, Methods in Molecular Biology 2nd edition (2009). Humana Press, Totowa NJ, Chapter 2, 3.
- Zrein M, Obert G, van Regenmortel MHV. Use of egg-yolk antibody for the detection of respiratory Syncytial virus in nasal secretions by ELISA. Arch Virol 90 (1986): 197-206.
- de Kloet E,de Kloet SR. Analysis of the beak and feather disease viral genome indicates the existence of several genotypes which have a complex psittacine host specificity. Arch Virol 149 (2004): 2393-2412.
- de Kloet SR. Sequence analysis of four double-stranded RNA genomic segments reveals an orthoreovirus with a unique genotype infecting psittaciformes. Avian Dis 52 (2008): 480-486.
- Marton S,Bányai K,Gál J, et al. Coding-complete sequencing classifies parrot bornavirus 5 into a novel virus species. Arch Virol 160 (2015): 2763-2768.
- Guo J, Tizard I. The genome sequence of parrot bornavirus 5. Virus Genes 51 (2015): 430-433.
- Sa-ardta P, Rinder M, Korbel R. First detection and characterization of Psittaciform bornaviruses in naturally infected and diseased birds in Thailand. Vet. Microbiol 230 (2019): 62-71.
- Komorizono R, Makino A, Horie M, et al. Sequence determination of a new parrotbornavirus-5strainin Japan: implications of clade-specific sequence diversity in the regions interacting with host factors. Microbiol Immunol 60 (2016): 437-441.
- Rubbenstroth D, Schlottau K, Schwemmle M, et al. Humanbornavirusresearch: Back on track! PLoS Pathog 15 (2019): e1007873.
- Zimmermann V,Rinder M,Kaspers B,et al.Impact of antigenic diversity on laboratory diagnosis of Avian bornavirus infections in birds. J Vet Diagn Invest 26 (2014): 769-777.
- Prager EM,Wilson AC. The dependence of immunological cross-reactivity upon sequence resemblance among lysozymes. I. Micro-complement fixation studies. J Biol Chem 246 (1971): 5978-5989.
- Prager EM. The sequence-immunology correlation revisited; Data for Cetacean Myoglobin and mammalian lysozymes. J Mol Evol 37 (1993): 408-416.
- Davidson I, Raibstein I, Altory-Natour A, et al. Development of duplex dual-gene and DIVA real-time RT-PCR assays and use of feathers as a non-invasive sampling method for diagnosis of Turkey Meningoencephalitis Virus. Avian Pathol 46 (2017): 256-264.