The Cancer Epigenome: A Review
Article Information
James A Marcum*
Baylor University, 1 Bear Pl #97273, Waco, TX, USA
*Corresponding Author: James A Marcum, Baylor University, 1 Bear Pl #97273, Waco, TX, USA
Received: 08 July 2019; Accepted: 08 August 2019; Published: 14 August 2019
Citation:
James A Marcum. The Cancer Epigenome: A Review. Journal of Biotechnology and Biomedicine 2 (2019): 067-083.
Share at FacebookAbstract
This review covers the current literature, since the completion of the Human Genome Project, on the cancer epigenome, including both cancer epigenetics and epigenomics. To that end, the transition from the human genome to the cancer genome is initially discussed, especially in terms of the hallmarks of cancer and its associated somatic mutation theory of cancer, along with the failure of this theory-based strictly on genetic mutations-to account for carcinogenesis and metastasis. The cancer epigenome (as well as both cancer epigenetics and epigenomics) is examined next, especially with respect to its role in tumorigenesis. The review’s main goal is to address what constitutes the cancer epigenome vis-à-vis the cancer genome and what the relationship is between them.
Keywords
Cancer, Carcinogenesis, Epigenetics, Epigenome, Epigenomics, Genome, Metastasis, Tumorigenesis
Cancer articles Cancer Research articles Cancer review articles Cancer PubMed articles Cancer PubMed Central articles Cancer 2023 articles Cancer 2024 articles Cancer Scopus articles Cancer impact factor journals Cancer Scopus journals Cancer PubMed journals Cancer medical journals Cancer free journals Cancer best journals Cancer top journals Cancer free medical journals Cancer famous journals Cancer Google Scholar indexed journals Carcinogenesis articles Carcinogenesis Research articles Carcinogenesis review articles Carcinogenesis PubMed articles Carcinogenesis PubMed Central articles Carcinogenesis 2023 articles Carcinogenesis 2024 articles Carcinogenesis Scopus articles Carcinogenesis impact factor journals Carcinogenesis Scopus journals Carcinogenesis PubMed journals Carcinogenesis medical journals Carcinogenesis free journals Carcinogenesis best journals Carcinogenesis top journals Carcinogenesis free medical journals Carcinogenesis famous journals Carcinogenesis Google Scholar indexed journals Epigenetics articles Epigenetics Research articles Epigenetics review articles Epigenetics PubMed articles Epigenetics PubMed Central articles Epigenetics 2023 articles Epigenetics 2024 articles Epigenetics Scopus articles Epigenetics impact factor journals Epigenetics Scopus journals Epigenetics PubMed journals Epigenetics medical journals Epigenetics free journals Epigenetics best journals Epigenetics top journals Epigenetics free medical journals Epigenetics famous journals Epigenetics Google Scholar indexed journals Epigenome articles Epigenome Research articles Epigenome review articles Epigenome PubMed articles Epigenome PubMed Central articles Epigenome 2023 articles Epigenome 2024 articles Epigenome Scopus articles Epigenome impact factor journals Epigenome Scopus journals Epigenome PubMed journals Epigenome medical journals Epigenome free journals Epigenome best journals Epigenome top journals Epigenome free medical journals Epigenome famous journals Epigenome Google Scholar indexed journals Genome articles Genome Research articles Genome review articles Genome PubMed articles Genome PubMed Central articles Genome 2023 articles Genome 2024 articles Genome Scopus articles Genome impact factor journals Genome Scopus journals Genome PubMed journals Genome medical journals Genome free journals Genome best journals Genome top journals Genome free medical journals Genome famous journals Genome Google Scholar indexed journals Epigenomics articles Epigenomics Research articles Epigenomics review articles Epigenomics PubMed articles Epigenomics PubMed Central articles Epigenomics 2023 articles Epigenomics 2024 articles Epigenomics Scopus articles Epigenomics impact factor journals Epigenomics Scopus journals Epigenomics PubMed journals Epigenomics medical journals Epigenomics free journals Epigenomics best journals Epigenomics top journals Epigenomics free medical journals Epigenomics famous journals Epigenomics Google Scholar indexed journals Metastasis articles Metastasis Research articles Metastasis review articles Metastasis PubMed articles Metastasis PubMed Central articles Metastasis 2023 articles Metastasis 2024 articles Metastasis Scopus articles Metastasis impact factor journals Metastasis Scopus journals Metastasis PubMed journals Metastasis medical journals Metastasis free journals Metastasis best journals Metastasis top journals Metastasis free medical journals Metastasis famous journals Metastasis Google Scholar indexed journals Tumorigenesis articles Tumorigenesis Research articles Tumorigenesis review articles Tumorigenesis PubMed articles Tumorigenesis PubMed Central articles Tumorigenesis 2023 articles Tumorigenesis 2024 articles Tumorigenesis Scopus articles Tumorigenesis impact factor journals Tumorigenesis Scopus journals Tumorigenesis PubMed journals Tumorigenesis medical journals Tumorigenesis free journals Tumorigenesis best journals Tumorigenesis top journals Tumorigenesis free medical journals Tumorigenesis famous journals Tumorigenesis Google Scholar indexed journals diagnosis articles diagnosis Research articles diagnosis review articles diagnosis PubMed articles diagnosis PubMed Central articles diagnosis 2023 articles diagnosis 2024 articles diagnosis Scopus articles diagnosis impact factor journals diagnosis Scopus journals diagnosis PubMed journals diagnosis medical journals diagnosis free journals diagnosis best journals diagnosis top journals diagnosis free medical journals diagnosis famous journals diagnosis Google Scholar indexed journals Cancer Genome articles Cancer Genome Research articles Cancer Genome review articles Cancer Genome PubMed articles Cancer Genome PubMed Central articles Cancer Genome 2023 articles Cancer Genome 2024 articles Cancer Genome Scopus articles Cancer Genome impact factor journals Cancer Genome Scopus journals Cancer Genome PubMed journals Cancer Genome medical journals Cancer Genome free journals Cancer Genome best journals Cancer Genome top journals Cancer Genome free medical journals Cancer Genome famous journals Cancer Genome Google Scholar indexed journals
Article Details
1. Introduction
Completion of the Human Genome Project in 2003 paved the way for the emergence of a systems biology approach to the investigation of complex living organisms and their associated diseases [1-3]. The current high-throughput omics assays, such as genomics, proteomics, and metabolomics, provide vast amounts of data concerning the dynamic states of cells and tissues, whether normal or pathological. Biomedical scientists today employ systems biology computational analyses and models to gain insights into high-throughput data via bioinformatics, especially for developing safe and effective therapeutic agents.
In this paper, the current literature on the cancer epigenome, as well as on cancer epigenetics and epigenomics, is reviewed. To that end, the transition from the human genome to the cancer genome is discussed initially, especially in terms of the hallmarks of cancer. The somatic mutation theory (SMT) of cancer is also discussed, along with its failure to account for carcinogenesis. Part of the reason for its failure is that the theory is based strictly on genetic mechanisms, particularly gene mutations [4]. Next, epigenetics and epigenomics are introduced and discussed in terms of the various molecular mechanisms involved in regulating gene activity. The epigenome is then examined in terms of its hierarchical structure of nucleosome, chromatosome, chromatin, and chromosome, and with respect to its role as the conceptual and empirical means for yoking an organism’s genotype with its phenotype. The discussion then turns to the cancer epigenome, including both cancer epigenetics and epigenomics, and to the role epigenetic mechanisms play in tumorigenesis. The review concludes with a summary statement of what constitutes the cancer epigenome and its reciprocal relationship to the cancer genome.
2. From the Genome to the Cancer Genome
In 1971, then U.S. President Richard Nixon declared war on cancer and assured the American public that the disease would be vanquished by its bicentennial year-1976 [5]. Unfortunately, that war has yet to be won and some have succumbed to the belief that the battle in terms of identifying some of the cancer genes may have been won, but the war with respect to translating these genes into actionable therapy has not [6]. But, optimism that the war can still be won is based on the belief that cancer is strictly a genetic disease and that recent technological advances, such as the development of next-generation DNA sequencing and various omics technologies, can not only identify the driver genes responsible for tumorigenesis but also elucidate the mechanism responsible for the origins of tumors and their capacity to invade other tissues within the body [7-10]. In the remainder of this section, the transition from of the human genome to the cancer genome is examined briefly, especially in terms of the hallmarks of cancer and the SMT of cancer. Although there are multiple definitions for the genome, the U.S. National Institutes of Health (NIH) provides a succinct but ample definition: the genome is an organism’s complete set of DNA, including all of its genes. Each genome contains all of the information needed to build and maintain that organism. In humans, a copy of the entire genome-more than 3 billion DNA base pairs-is contained in all cells that have a nucleus [https://ghr.nlm.nih.gov/primer/hgp/genome].
In the early 1990s, the NIH, along with other international agencies initiated the Human Genome Project to sequence the human genome [11-13]. The project was officially completed in 2003, which marshalled in the postgenomic era and the different high-throughput omics technologies associated with it [14-16]. Basically, the shift from the genomic era to the postgenomic era reflected the ability to obtain high-throughput omics data not just on a single person but also on a population and not simply from a single time-point but also from multiple time points. In other words, biologists, especially systems biologists, could now investigate living processes as dynamic processes. Finally, one of the more interestingly immediate outcomes of the postgenomic era is the observation of incredible variation among genomic expression, even within the same cells of a specific tissue, prompting one reviewer to acknowledge that no “single genome” exists [17].
The postgenomic era has had a tremendous impact on medicine and on the diagnosis and treatment of diseases [18-20]. This impact has been especially realized in cancer biology, with several initiatives supporting projects to elucidate the cancer genome [21, 22]. One of the more prominent projects is the Cancer Genome Project, which was launched in 2000 under the aegis of the U.K. Wellcome Trust Sanger Institute [23, 24]. The project is also associated with the International Cancer Genome Consortium (ICGC), which was assembled in 2008 and supports international research on the cancer genome [25]. Another project is the NIH’s The Cancer Genome Atlas (TCGA), which was announced in 2005 and several pilot projects were funded the following year [26]. The project was officially completed in 2015, with the discovery of over ten million cancer-associated mutations [27].
One of the main goals of the TCGA project was to identify the genetic mutations responsible for the hallmarks of cancer, especially as articulated in the SMT of cancer [28, 29]. The first hallmark is the cancer cell’s self-sufficiency of endogenous growth factors, such as activated oncogenes like MYC and RAS. The next hallmark is the flip side of the first in terms of the cancer cell’s insensitivity to antigrowth signals, such as tumor suppressor genes like p53 and PTEN. These two hallmarks converge with respect to a third-the cancer cell’s limitless replicative potential, i.e. cellular immortality. Associated with these hallmarks is the hallmark of altered metabolism or the Warburg effect, i.e. cancer cells favor glycolysis. A fifth hallmark associated with the previous four hallmarks is the cancer cell’s ability to evade cell death or apoptosis. Besides evading cell death, the cancer cell can also evade detection by the immune system. Collectively, these hallmarks allow for growth but limited by the diffusion of nutrients and waste products. Substantial growth begins with the next hallmark, angiogenesis or the recruitment of a blood vessels to the tumor. The eighth hallmark is local chronic inflammation, which can accelerate angiogenesis and induce an associated remodeling of the extracellular matrix that leads to the penultimate hallmark-metastasis, which involves the cancer cell’s invasion of other tissues. The metastasis hallmark is claimed to be the true hallmark of malignant tumors as compared to benign tumors [30]. The final hallmark is genomic instability, which can manifest itself with respect to genome aneuploidy and chromosomal rearrangement. Conceptually, these hallmarks and the SMT of cancer have been very influential is guiding cancer research [31, 32], although they have been criticized, especially with respect to not providing sufficient understanding of tumorigenic mechanisms for the development of effective therapeutic drugs [33, 34].
3. From the Epigenome to the Cancer Epigenome
Part of the problem with the hallmarks of cancer and its associated the SMT of cancer is that they emphasize mutations of oncogenes and tumor suppressor genes. Unfortunately, these mutations have a limited impact on phenotypic traits of cancer cells and tumors [35]. Although proponents of the SMT acknowledge that epigenetic mechanisms are involved in genetic regulation, e.g. the final hallmark of cancer, these mechanisms are not completely developed with respect to carcinogenesis and metastasis. In this section, epigenetic and epigenomic mechanisms are discussed first, followed by an examination of the hierarchical structure of the epigenome in terms of the nucleosome, chromatosome, chromatin, and chromosome. The technologies involved in epigenomics are also introduced and discussed, before turning to the cancer epigenome and its associated epigenetics and epigenomics. The section’s chief goal is to bring conceptual clarity to what is meant by the epigenome, as well as by epigenetics and epigenomics, especially since there are no consensus definitions for these terms.
In 1942, Conrad Waddington [36] introduced the term epigenetics, along with the notion of epigenotype, to bridge the gap between genotype and phenotype. His goal was to provide a genetic basis for embryological and morphological development. In other words, epigenetics conceptually is the progeny of wedding the historical embryological theory of epigenesis with Mendelian genetics (epi + genetics) [37]. Waddington [38] later introduced the epigenetic landscape to illustrate the connection between the genotype and phenotype in that the genes constituting the genome shape the phenotypic landscape as the zygote or cell proceeds towards its differentiated state.
With the advent of molecular biology, epigenetics took a molecular turn from its previous morphologic conception [39-42]. Although there are multiple definitions for epigenetics in the literature, with each emphasizing different dimensions of the term [43-46], Carrie Deans and Keith Maggert propose the following definition to capture these various dimensions. They define epigenetics as “the study of phenomena and mechanisms that cause chromosome-bound, heritable changes to gene expression that are not dependent on changes to DNA sequence [47, p. 892]. The definition tethers the changes in gene expression to chromosomes independent of their DNA sequence or composition and it acknowledges that these changes are heritable. The term phenomena is broad enough to include not only developmental or embryological processes but also other biological and pathological processes, such as tumorigenesis. Finally, the term mechanism can refer to the molecular and morphological dimensions of epigenetics.
As illustrated in Figure 1, the molecular mechanisms comprising contemporary epigenetics can be divided into at least three major categories [48-50]. The first is DNA methylation in which a methyl group is transferred to cytosine at its 5-carbon position, generating 5-methylcytosine [51-53]. The cytosine generally precedes guanosine, forming a CpG dinucleotide. CpG-rich areas of the genome are called CpG islands and are often associated with promoter regions of genes. Methylation generally silences a gene by prohibiting its transcription. The profiling of DNA methylation patterns is called the methylome [54] and has been mapped [55], as well as used in concert with transcriptome profiling to investigate and define the larger epigenetic landscape [56]. Finally, DNA methylation is reversible with the removal of the methyl group.
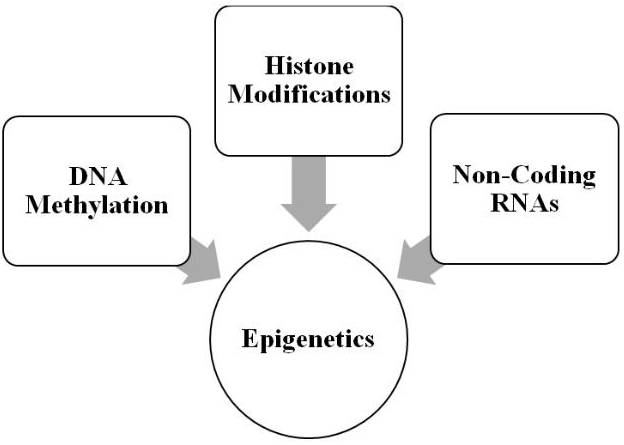
Figure 1: Molecular mechanisms comprising epigenetics (see text for details).
The next category of epigenetic mechanisms consists of the various histone post-translational modifications (PTMs), which include acetylation, methylation, and phosphorylation-to list the best studied PTMs-and importantly the modifications are also reversible [57-59]. The histone proteins are divided into five basic types: H1, H2A, H2B, H3, and H4. These proteins are important for packaging the DNA; and, histone modifications are responsible for altering chromatin architecture. Thus, PTMs alter the state of the packaging from a tightly wound state (heterochromatin), which inhibits transcription, to a relaxed state (euchromatin), which promotes it. Currently, the mechanism for PTMs is articulated in terms of “writers” or the enzymes that modify the histones, “erasers” or the enzymes that remove the modifications, and “readers” or the proteins that recognize modification sites on the histone and are involved in interpreting them with respect to downstream processes [60, 61]. Finally, there are several databases available for storing and curating histone PTMs [62, 63].
The last category of epigenetic mechanisms consists of non-coding RNAs (ncRNAs), which are transcribed from DNA but are not translated into protein; and, these ncRNAs are distinct from messenger, ribosomal, and transfer RNAs [64-67]. In general, ncRNAs range in epigenetic functions from DNA methylation and gene inactivation to chromatin remodeling. They are divided into two major groups: short ncRNAs, which are <30 nucleotides in length, and long ncRNAs, which are >200 nucleotides in length. Short ncRNAs are divided into several types: microRNAs, which bind messenger RNA and prevent its translation, short-interfering RNAs, which share a similar function to microRNAs and also induce heterochromatin formation, PIWI-interacting RNAs, which bind the PIWI proteins and suppress transposon activity, and small nucleolar RNAs (these ncRNAs are intermediate in size compared to short and long ncRNAs), which are involved regulating translation through ribosomal activity. Long ncRNA is the largest group of ncRNAs and are generally involved in chromatin remodeling and other molecular mechanisms. Two important types are enhancer RNAs and circular RNAs, which are involved in transcriptional regulation.
In sum, these epigenetic mechanisms provide the molecular foundation for conceptualizing the epigenome, which is made possible by a shift from epigenetics and single gene studies to epigenomics and epigenome-wide association studies, as well as various high-throughput assays and next-generation sequencing [68-70]. One of the earliest epigenomic technologies was combining DNA microarrays with chromatin immunoprecipitation technology (ChIP-Chip), which has been used to determine the global pattern of histone PTMs. Today, ChIP-Seq incorporates next-generation sequencing to provide higher resolution of histone modifications. Other technologies include bisulfite and nanopore sequencing and chromatin accessibility and interaction technologies. Finally, several consortia have been initiated to facilitate collaborative epigenomic research, including ECODE (Encyclopedia of DNA Elements), FANTOM (Functional Annotation of the Mouse/Mammalian Genome), and the Roadmap Epigenomics Consortium [71].
While the genome represents the linear DNA sequence of the genes, especially those coding for proteins, which are believed responsible for phenotypic traits, the epigenome pertains to the structural elements composed of both DNA and proteins that constitute the three-dimensional (3D) patterns or markings that constitute the landscape ultimately responsible for regulating gene expression [72]. And, when the Human Genome Project was completed in 2003 the
Human Epigenome Project was initiated [73], followed later by the launching of the International Human Epigenome Consortium [74]. Moreover, recent technological advances, such as single cell epigenome sequencing and CRISPR editing of the epigenome [76], are providing further insights into and use of the epigenome. As depicted in Figure 2, four structural elements, the nucleosome, chromatosome, chromatin, and chromosome, comprise the epigenomic hierarchy; and, the structural state of these elements contains the epigenomic information necessary for regulating gene and transcription activity.
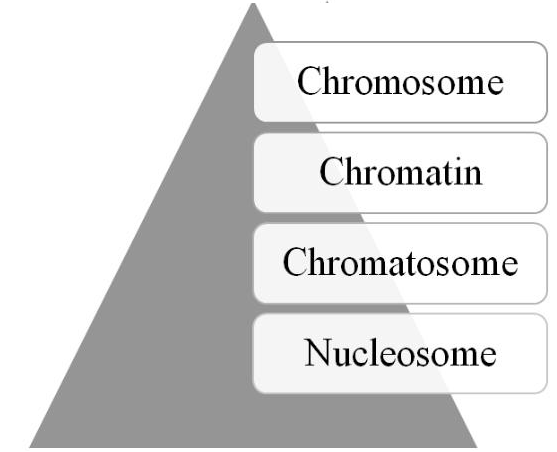
Figure 2: The epigenome’s structural hierarchy (see text for details).
The basic structural unit of the epigenome is the nucleosome [77-79]. The nucleosome is composed of DNA strands (~146 nucleotides) wrapped ~1.6 turns around an octamer of core histone proteins (2 each of H2A, H2B, H3, and H4). Both the histone’s N-terminal tails and the lateral surface of globular bodies contain sites or marks for PTMs. A sequence of relaxed nucleosomes are ~200 nucleotides apart and resemble beads on a string. The next structure is the chromatosome in which the histone protein H1 or linker histone, along with other non-histone proteins, binds via linker DNA to the nucleosome [80-82]. The chromatosome consists of ~166 nucleotides and has a diameter of ~10 nm. It is the basic unit involved in the folding process to form chromatin. The process depends on the epigenetic information, called the histone code [83, 84], which is contained in the chromatosome, especially on its histone tails. The code consists of PTMs patterns that have an impact on the folding process of larger epigenomic structures. Although there is some debate as to whether it is a code or language, there is general consensus that it and the crosstalk among the histone PTMs, as well as DNA modifications, function as the molecular mechanism for generating the epigenome’s next hierarchical structural element-chromatin [85-87].
Traditionally, chromatin is defined operationally as the fibers or granules in the nucleus that are observed after staining [88]. It can be compacted at various levels, ranging up to 20,000 fold, generating what are called “higher-order” structures [89-91]. Indeed, the epigenome contains sufficient information for structuring the chromatin landscape in terms of various domains that they are involved in gene regulation [92, 93]. In short, chromatin is the fundamental structural unit that composes the final and largest structural element of the epigenome—chromosomes [94].
Chromosomes are defined historically as rod-like structures within the cell’s nucleus that contain the genetic information for heritable traits, i.e. the chromosome theory of inheritance [95, 96]. But, chromosomes also carry epigenomic information based on their architecture, which is critical for gene regulation. For example, experimental evidence has been recently reported to suggest that epigenetic marking patterns contain sufficient information for determining a chromosome’s global 3D architecture [97]. Finally, during cellular interphase, chromosome are generally in a relaxed or euchromatin state, which can be divided into territories [98-100]. These territories contain channels that are important not simply in regulating gene expression but also in optimizing it.
As the epigenome consists of the normal architecture of chromosomal structure for the transmission of epigenetic information, the cancer epigenome consists of the abnormal chromosomal architecture that results in a distortion of epigenetic information [101-103]. The cancer epigenome “harbors myriad abnormalities that are based on somatically heritable alterations that are not due to primary DNA sequence changes” [104, p. 726]. One of the chief hallmarks of the cancer epigenome is chromosomal instability (CIN), which is the result of genomic instability-especially with mutation to DNA repair genes and other mitotic regulatory elements, such as chromatin remodeler complexes [105, 106]. Specifically, CIN is caused by mis-segregation of chromosomes during mitosis. It can be classified as either structural CIN or numerical CIN [107]. Structural CIN is the alteration of chromosomal architecture, such as loss of chromosomal material in such processes as chromothripsis. Numerical CIN involves abnormal chromosomal number, such as aneuploidy or polyploidy. CIN is found in well over half of cancer patients (60-80%) and it is associated with most stages of carcinogenesis, including metastasis [110]. Moreover, both structural and numerical CIN do not necessarily occur together in every cancer [108, 109]. Finally, chromosome territories are also involved in tumorigenesis through their rearrangement so genes that are often insulated from one another intermingle [111-113].
Chromatin architecture and remodeling, especially in terms of chromatin remodeler complexes, play an important role in carcinogenesis and metastasis [114-116]. As noted earlier, chromatin has two major configurations that have an impact on gene activity through access of transcription factors. The first is the relaxed or permissive configuration (euchromatin) in which there is an increase in histone PTMs, such as H3K4me3 and H3K27ac. The net effect is to lower the barriers of the epigenetic landscape, i.e. increase the spacing among nucleosomes, to permit cell state transitions. The second chromatin configuration is compacted or restrictive (heterochromatin) in which there is generally a decrease in histone PTMs, such as deacetylation or demethylation, with the overall effect of heightening the epigenetic barriers, i.e. decrease the spacing among nucleosome, and thereby prohibiting cell state transitions. Moreover, other histone PTMs are associated with carcinogenesis. For example, phosphorylation of the linker histone H1 at threonine 146 has been found in various breast cancer cell lines [117]. Finally, the histone code has been proposed to play a role in tumorigenesis, in terms of both the epigenetic factors shaping it and the code’s role in directing the activity of remodeler chromatin complexes [118, 119]-although mechanistic details remain unclear.
Besides histone PTMs, the other two epigenetic mechanisms depicted in Figure 1 also play a role in carcinogenesis and metastasis [120-123]. There are two patterns of DNA methylation in tumorigenesis that make up the cancer methylome [124, 125]. The first pattern or methylome involves DNA hypermethylation, especially of CpG islands associated with the promoter regions of tumor suppressor genes. For example, the tumor suppressor gene p16INK4a is commonly hypermethylated in breast cancer and its silencing has an impact on remodeling the chromatin boundary, which contributes to tumorigenesis [126]. Besides hypermethylation, global DNA or genome-wide hypomethylation, particularly of proto-oncogenes and growth factors, plays a key role in carcinogenesis. For example, global demethylation in NIH3T3 cells is associated with their oncogenic transformation [127]. Besides CpG islands, altered methylation patterns are also associated with CpG regions distal (~2 kilobases) to the islands or what are called CpG shores. For example, colon cancer includes not only hypermethylation of CpG islands but also hypomethylation of the CpG shores [128]. In sum, changes in methylation patterns or methylomes significantly alter epigenetic integrity and leads to carcinogenesis and metastasis.
The final epigenetic molecular mechanism in tumorigenesis is ncRNA [129-132]. As for the small ncRNAs, microRNAs represent an important class for epigenetic regulation. For example, the microRNA, miR-132, which plays an antimetastatic role through controlling cell adhesion, is downregulated in prostate cancer via CpG island hypermethylation [133]. The microRNAs miR-106b, miR-25, miR-93, miR-23a and miR-27a, which are upregulated in hepatocellular carcinoma via hypomethylation of CpG poor regions or shores, are another example [134]. Long ncRNAs also play a major role in carcinogenesis. For example, growth arrest-specific transcript 5, which controls cellular apoptosis, is a long ncRNA, whose gene GAS5 is downregulated in breast cancer [135]. Another important example is the long ncRNA, HOTAIR, which is also associated not only with breast cancer but also with gastric, colorectal, ovarian, and lung cancers; and, it is involved in altering chromatin dynamics through its interaction with the histone methyltransferase Polycomb Repressive Complex 2 and lysine-specific demethylase 1 [136, 137]. Indeed, given the diverse function of long ncRNAs, ranging from gene regulation to chromatin restructuring and dynamics, long ncRNAs are making a significant impact on cancer biology and are responsible for a paradigm shift from the traditional paradigm based exclusively on protein-coding RNA [138, 139].
Finally, just as genetic changes can account for the hallmarks of cancer, so too can epigenetic changes [140-144]. For example, the methylation of the RASSF1A (RAS association domain-containing protein 1) gene, in both lung and breast cancers, results in increased RAS signaling and thereby contributes to the hallmark of the cancer cell’s self-sufficiency of endogenous growth factors [145]. Another example is the downregulation of the DAB2 (disabled homolog 2 protein) gene through methylation, which converts TGF-β (transforming growth factor-β) to a potent oncogenic factor in human squamous cell carcinomas and thereby accounts for the hallmark of insensitivity to antigrowth signals [146]. Another example is the silencing of the p16 gene through promoter methylation in non?small cell lung cancers and is consequently responsible for a cancer cell’s limitless replicative potential [147]. An epigenetic example for the hallmark of evading cell death or apoptosis is the methylation of the HIC1 (Hypermethylated in cancer 1 protein) gene, which is involved in regulating cell proliferation and apoptosis in prostate cancer [148]. An example of the angiogenesis hallmark is the downregulation of the anti-angiogenic genes, TIMP3 (tissue inhibitor of metalloproteinase 3) and CDH1 (E-cadherin), in ovarian cancer through DNA methylation and histone modification [149]. A final example pertains to the metastasis hallmark in which methylated silencing of the gene for the microRNA, miR-148a, in primary tumors results in the upregulation of oncogenic and metastatic genes [150].
4. Conclusion
Conceptually, the epigenome pertains to the epigenetic information stored in the packaging of the epigenome. This information, which consists of epigenetic marks, is responsible for the genome’s expression and for a cell’s transition to its differentiated state. Whereas the genome consists of a linear sequence of genes that contain the code for transcription products, the epigenome consists of a 3D configuration of marks that compose a code for regulating the expression of those products. While the genome is responsible for the genotype, the epigenome serves as the intermediary or mediating mechanism for interpreting the genotype qua phenotype. In short, the epigenome is the link between genotype and phenotype. Finally, the relationship between the genome and epigenome is reciprocal (Figure 3). Thus, changes in the genome can have an impact on the epigenome and vice versa, which has important implications for elucidating carcinogenesis and metastasis.
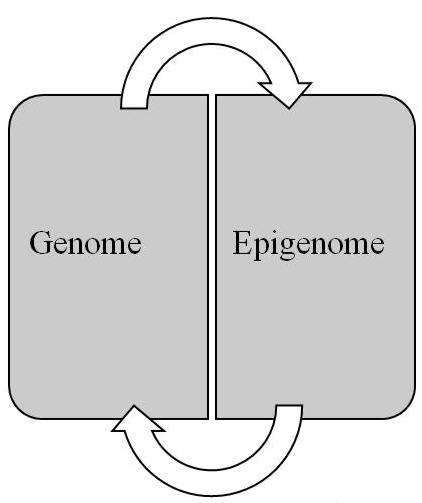
Figure 3: Reciprocal relationship between genome and epigenome (see text for details).
Although the SMT of cancer emphasizes mutations in oncogenes and tumor suppressor genes (the first two hallmarks of cancer) for cellular oncogenic transformation, the initial mutations probably do not involve these genes but rather genes for chromatin remodelers [151], for example, which alter chromatin’s architecture and thereby its epigenetic information. Given the reciprocal relationship between the genome and epigenome (Figure 3), genetic mutations to such genes initially lead to the cancer epigenome, which then contributes to an altered expression of the genome that results in fulfilment of the hallmarks of cancer. Moreover, just as the environment can cause genetic mutations so it can also cause epigenetic mutations, i.e. changes in the epigenome’s architecture, which can alter the genome’s expression. In sum, as depicted in Figure 4, carcinogenesis and metastasis are the outcome of an imbalance in mitotic homeostasis caused by oncogenic changes in both the genome and epigenome, with priority given initially to changes in the epigenome. Finally, treatment of cancer must take both the genome and epigenome into consideration to develop accurate diagnostic protocols and efficacious therapy.
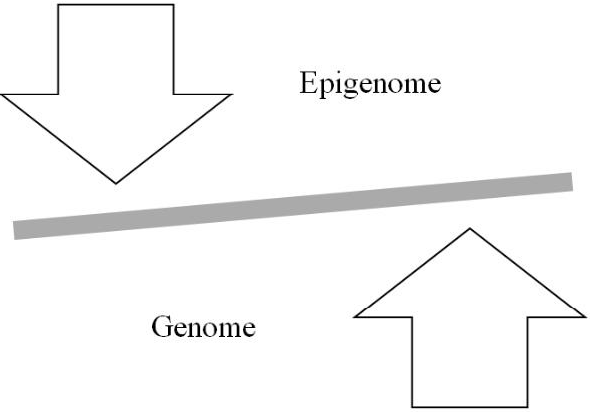
Figure 4: Imbalance in mitotic homeostasis (see text for details).
Conflict of Interest
The author declares no conflict of interest.
References
- Hood L. Systems Biology: Integrating Technology, Biology, and Computation. Mechanisms of Ageing and Development 124 (2003): 9-16.
- Check Hayden E. Human Genome at Ten: Life is Complicated. Nature News 464 (2010): 664-667.
- Hood L, Rowen L. The Human Genome Project: Big Science Transforms Biology and Medicine. Genome Medicine 5 (2013): 79.
- Soto AM, Sonnenschein The Somatic Mutation Theory of Cancer: Growing Problems with the Paradigm? BioEssays 26 (2004): 1097-1107.
- Sporn MB. The War on Cancer: A Review. Annals of the New York Academy of Sciences 833 (1997): 137-146.
- Brennan R, Federico S, Dyer MA. The War on Cancer: Have We Won the Battle but Lost the War? Oncotarget 1 (2010): 77-83.
- Hayes DN, Kim WY. The Next Steps in Next-gen Sequencing of Cancer Genomes. Journal of Clinical Investigation 125 (2015): 462-468.
- Kruger R. Charting a Course to a Cure. Cell 173 (2018): 277.
- Patel NM, Michelini VV, Snell JM, et al. Enhancing Next?Generation Sequencing?Guided Cancer Care through Cognitive Computing. Oncologist 23 (2018): 179-185.
- Breitenbach M, Hoffmann J. Cancer Models 2018. Frontiers in Oncology 8 (2018): 401.
- Watson JD. The Human Genome Project: Past, Present, and Future. Science 248 (1990): 44-49.
- Cantor CR. Orchestrating the Human Genome Project. Science 248 (1990): 49-51.
- Collins FS, Patrinos A, Jordan E, et al. New Goals for the US Human Genome Project: 1998-2003. Science 282 (1998): 682-689.
- Collins FS, Morgan M, Patrinos A. The Human Genome Project: Lessons from Large-scale Biology. Science 300 (2003): 286-290.
- Nerlich B, Hellsten I. Genomics: Shifts in Metaphorical Landscape between 2000 and 2003. New Genetics and Society 23 (2004): 255-268.
- Kandpal RP, Saviola B, Felton J. The Era of ’Omics Unlimited. BioTechniques 46 (2009): 351-355.
- Falconer E. Looking Beyond the Post-Genomic Era. Genome Biology 14 (2013): 313.
- Peltonen L, McKusick VA. Dissecting Human Disease in the Postgenomic Era. Science 291 (2001): 1224-1229.
- Goh KI, Cusick ME, Valle D, et al. The Human Disease Network. Proceedings of the National Academy of Sciences, USA 104 (2007): 8685-8690.
- Loscalzo J, Kohane I, Barabasi AL. Human Disease Classification in the Postgenomic Era: A Complex Systems Approach to Human Pathobiology. Molecular Systems Biology 3 (2007): 124.
- Wheeler DA, Wang L. From Human Genome to Cancer Genome: The First Decade. Genome Research 23 (2013): 1054-1062.
- Ledford H. Big Science: The Cancer Genome Challenge. Nature 464 (2010): 972-974.
- Dickson D. Wellcome Funds Cancer Database. Nature 401 (1999): 729.
- Stratton MR, Campbell PJ, Futreal PA. The Cancer Genome. Nature 458 (2009): 719-724.
- International Cancer Genome Consortium. International Network of Cancer Genome Projects. Nature 464 (2010): 993-998.
- Collins FS, Barker AD. Mapping the Cancer Genome. Scientific American 296 (2007): 50-57.
- Ledford H. End of Cancer-Genome Project Prompts Rethink. Nature 517 (2015): 128129.
- Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell 100 (2000): 57-70.
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell 144 (2011): 646-674.
- Lazebnik Y. What are the Hallmarks of Cancer? Nature Reviews Cancer 10 (2010): 232-233.
- Fouad YA, Aanei C. Revisiting the Hallmarks of Cancer. American Journal of Cancer Research 7 (2017): 1016-1036.
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research 47 (2018): 941-947.
- Floor SL, Dumont JE, Maenhaut C, et al. Hallmarks of Cancer: Of All Cancer Cells, All the Time? Trends in Molecular Medicine 18 (2012): 509-515.
- Sonnenschein C, Soto AM. The Aging of the 2000 and 2011 Hallmarks of Cancer Reviews: A Critique. Journal of Biosciences 38 (2013): 651-663.
- Marcum JA. Cancer: Complexity, Causation, and Systems Biology. Medicina and Storia 9 (2009): 267-287.
- Waddington CH. The Epigenotype. Endeavour 1 (1942): 18-20.
- Lappalainen T, Greally JM. Associating Cellular Epigenetic Models with Human Phenotypes. Nature Reviews Genetics 18 (2017): 441-451.
- Waddington CH. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. (1957): London, Allen and Unwin.
- Jablonka E, Lamb MJ. The Changing Concept of Epigenetics. Annals of the New York Academy of Sciences 981 (2002): 82-96.
- Haig D. The (Dual) Origins of Epigenetics. Cold Spring Harbor Symposia on Quantitative Biology 69 (2004): 67-70.
- Holliday R. Epigenetics: A Historical Overview. Epigenetics 1 (2006): 76-80.
- Deichmann U. Epigenetics: The Origins and Evolution of a Fashionable Topic. Developmental Biology 416 (2016): 249-254.
- Bird A. Perceptions of Epigenetics. Nature 447 (2007): 396-398.
- Berger SL, Kouzarides T, Shiekhattar R, et al. An Operational Definition of Epigenetics. Genes and Development 23 (2009): 781-783.
- Pisco AO, d’Herouel AF, Huang S. Conceptual Confusion: The Case of Epigenetics. BioRxiv (2016): 053009.
- Nicoglou A, Merlin F. Epigenetics: A Way to Bridge the Gap between Biological Fields. Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences 66 (2017): 73-82.
- Deans C, Maggert KA. What do You Mean, “Epigenetic”? Genetics 199 (2015): 887-896.
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A Landscape Takes Shape. Cell 128 (2007): 635-638.
- Korkmaz A, Manchester LC, Topal T, et al. Epigenetic Mechanisms in Human Physiology and Diseases. Journal of Experimental and Integrative Medicine 1 (2011): 139-147.
- Allis CD, Jenuwein T. The Molecular Hallmarks of Epigenetic Control. Nature Reviews Genetics 17 (2016): 487-500.
- Bird A. DNA Methylation Patterns and Epigenetic Memory. Genes and Development 16 (2002): 6-21.
- Jin B, Li Y, Robertson KD. DNA Methylation: Superior or Subordinate in the Epigenetic Hierarchy? Genes and Cancer 2 (2011): 607-617.
- Edwards JR, Yarychkivska O, Boulard M, et al. DNA Methylation and DNA Methyltransferases. Epigenetics and Chromatin 10 (2017): 23.
- Pelizzola M, Ecker JR. The DNA Methylome. FEBS Letters 585 (2011): 1994-2000.
- Wilson IM, Davies JJ, Weber M, et al. Epigenomics: Mapping the Methylome. Cell Cycle 5 (2006): 155-158.
- Hu Y, An Q, Guo Y, et al. Simultaneous Profiling of mRNA Transcriptome and DNA Methylome from a Single Cell. Methods in Molecular Biology 1979 (2019): 363-377.
- Bartova E, Krejci J, Harnicarova A, et al. Histone Modifications and Nuclear Architecture: A Review. Journal of Histochemistry and Cytochemistry 56 (2008): 711-721.
- Bannister AJ, Kouzarides T. Regulation of Chromatin by Histone Modifications. Cell Research 21 (2011): 381-395.
- Stillman B. Histone Modifications: Insights into Their Influence on Gene Expression. Cell 175 (2018): 6-9.
- Nicholson TB, Veland N, Chen T. Writers, Readers, and Erasers of Epigenetic Marks. In Eds.: Gray SG. Epigenetic Cancer Therapy, Amsterdam, Academic Press (2015): 31-66.
- Vaijayanthi T, Pandian GN, Sugiyama H. Chemical Control System of Epigenetics. Chemical Record 18 (2018): 1833-1853.
- Khare SP, Habib F, Sharma R, et al. HIstome-A Relational Knowledgebase of Human Histone Proteins and Histone Modifying Enzymes. Nucleic Acids Research 40 (2011): 337-342.
- Draizen EJ, Shaytan AK, Marino-Ramirez L, et al. HistoneDB 2.0: A Histone Database with Variants-an Integrated Resource to Explore Histones and Their Variants. Database (2016): 014.
- Costa FF. Non?Coding RNAs: Meet Thy Masters. BioEssays 32 (2010): 599-608.
- Kaikkonen MU, Lam MT, Glass CK. Non-Coding RNAs as Regulators of Gene Expression and Epigenetics. Cardiovascular Research 90 (2011): 430-440.
- Liu N, Pan T. RNA Epigenetics. Translational Research 165 (2015): 28-35.
- Wei JW, Huang K, Yang C, et al. Non-Coding RNAs as Regulators in Epigenetics. Oncology Reports 37 (2017): 3-9.
- Callinan PA, Feinberg AP. The Emerging Science of Epigenomics. Human Molecular Genetics 15 (2006): R95-R101.
- Rakyan VK, Down TA, Balding DJ, et al. Epigenome-Wide Association Studies for Common Human Diseases. Nature Reviews Genetics 12 (2011): 529-541.
- Flanagan Epigenome-Wide Association Studies (EWAS): Past, Present, and Future. Methods in Molecular Biology 1238 (2015): 51-63).
- Carlberg C, Molnár F. Human Epigenomics. (2018): New York, Springer. Bernstein BE, Meissner A, Lander ES. The Mammalian Epigenome. Cell 128 (2007): 669-681.
- Bernstein BE, Meissner A, Lander ES. The Mammalian Epigenome. Cell 128 (2007): 669-681
- Bradbury J. Human Epigenome Project-Up and Running. PLoS Biology 1 (2003): e82.
- Abbott A. Europe to Map the Human Epigenome. Nature 477 (2011): 518.
- Wen L, Tang F. Single Cell Epigenome Sequencing Technologies. Molecular Aspects of Medicine 59 (2018): 62-69.
- Holtzman L, Gersbach CA. Editing the Epigenome: Reshaping the Genomic Landscape. Annual Review of Genomics and Human Genetics 19 (2018): 43-71.
- Kornberg RD, Lorch Y. Twenty-five Years of the Nucleosome, Fundamental Particle of the Eukaryote Chromosome. Cell 98 (1999): 285-294.
- McGinty RK, Tan S. Nucleosome Structure and Function. Chemical Reviews 115 (2014): 2255-2273.
- Lawrence M, Daujat S, Schneider Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends in Genetics 32 (2016): 42-56.
- Simpson RT. Structure of the Chromatosome, a Chromatin Particle Containing 160 Base Pairs of DNA and All the Histones. Biochemistry 17 (1978): 5524-5531.
- Bharath MS, Chandra NR, Rao MR. Molecular Modeling of the Chromatosome Particle. Nucleic Acids Research 31 (2003): 4264-4274.
- Ozturk MA, Cojocaru V, Wade RC. Toward an Ensemble View of Chromatosome Structure: A Paradigm Shift from One to Many. Structure 26 (2018): 1050-1057.
- Jenuwein T, Allis CD. Translating the histone code. Science 293 (2001): 1074-1080.
- Gardner KE, Allis CD, Strahl BD. Operating on Chromatin, A Colorful Language Where Context Matters. Journal of Molecular Biology 409 (2011): 36-46.
- Lee JS, Smith E, Shilatifard A. The Language of Histone Crosstalk. Cell 142 (2010): 682-685.
- Rothbart SB, Strahl BD. Interpreting the Language of Histone and DNA Modifications. Biochimica et Biophysica Acta 1839 (2014): 627-643.
- Su Z, Denu JM. Reading the Combinatorial Histone Language. ACS Chemical Biology 11 (2015): 564-574.
- Van Holde KE. Chromatin. New York, Springer-Verlag (1989).
- Woodcock CL, Ghosh RP. Chromatin Higher-Order Structure and Dynamics. Cold Spring. Harbor Perspectives in Biology 2 (2010): a000596.
- Di Pierro M, Zhang B, Aiden EL, et al. Transferable Model for Chromosome Architecture. Proceedings of the National Academy of Sciences, USA 113 (2016): 12168-12173.
- Ghosh SK, Jost D. How Epigenome Drives Chromatin Folding and Dynamics, Insights from Efficient Coarse-Grained Models of Chromosomes. PLoS Computational Biology 14 (2018): e1006159.
- De Graaf CA, van Steensel B. Chromatin Organization: Form to Function. Current Opinion in Genetics and Development 23 (2013): 185-190.
- Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Molecular Cell 62 (2016): 668-680.
- Appels R, Morris R, Gill BS, et al. Chromosome Biology. (2012): New York, Springer Science and Business Media.
- Sutton WS. The Chromosomes in Heredity. Biological Bulletin 4 (1903): 231-250.
- Morgan TH. Chromosomes and Heredity. American Naturalist 44 (1910): 449-496.
- Di Pierro M, Cheng RR, Aiden EL, et al. De Novo Prediction of Human Chromosome Structures: Epigenetic Marking Patterns Encode Genome Architecture. Proceedings of the National Academy of Sciences, USA 114 (2017): 12126-12131.
- Meaburn KJ, Misteli T. Chromosome Territories 445 (2007): 379-381.
- Cremer T, Cremer M. Chromosome Territories. Cold Spring Harbor Perspectives in Biology 2 (2010): a003889.
- Fritz AJ, Sehgal N, Pliss A, et al. Chromosome Territories and the Global Regulation of the Genome. Genes Chromosomes and Cancer 58 (2019): 407-426.
- Ting AH, McGarvey KM, Baylin SB. The Cancer Epigenome-Components and Functional Correlates. Genes and Development 20 (2006): 3215-3231.
- Lechner M, Boshoff C, Beck S. Cancer Epigenome. Advances in Genetics 70 (2010): 247-276.
- Dawson MA. The Cancer Epigenome: Concepts, Challenges, and Therapeutic Opportunities. Science 355 (2017): 1147-1152.
- Baylin SB, Jones PA. A Decade of Exploring the Cancer Epigenome-Biological and Translational Implications. Nature Reviews Cancer 11 (2011): 726-734.
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic Instability-an Evolving Hallmark of Cancer. Nature Reviews Molecular Cell Biology 11 (2010): 220-228.
- Bakhoum SF, Landau DA. Chromosomal Instability as a Driver of Tumor Heterogeneity and Evolution. Cold Spring Harbor Perspectives in Medicine 7 (2017): a029611.
- Vargas-Rondon N, Villegas V, Rondon-Lagos M. The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 10 (2017): 4.
- Bakhoum SF, Cantley LC. The Multifaceted Role of Chromosomal Instability in Cancer and its Microenvironment. Cell 174 (2018): 1347-1360.
- Giam M, Rancati G. Aneuploidy and Chromosomal Instability in Cancer: A Jackpot to Chaos. Cell division 10 (2015): 3.
- Van Jaarsveld RH, Kops GJ. Difference Makers: Chromosomal Instability versus Aneuploidy in Cancer. Trends in Cancer 2 (2016): 561-571.
- Hnisz D, Schuijers J, Li CH, et al. Regulation and Dysregulation of Chromosome Structure in Cancer. Annual Review of Cancer Biology 2 (2018): 21-40.
- Umlauf D, Mourad R. The 3D Genome: From Fundamental Principles to Disease and Cancer. Seminars in Cell and Developmental Biology 90 (2018): 128-137.
- Szczepinska T, Rusek AM, Plewczynski D. Intermingling of chromosome territories. Genes Chromosomes and Cancer 58 (2019) Jan 21.
- Ellis L, Atadja PW, Johnstone RW. Epigenetics in Cancer: Targeting Chromatin Modifications. Molecular Cancer Therapeutics 8 (2009): 1409-1420.
- Nair SS, Kumar R. Chromatin Remodeling in Cancer: A Gateway to Regulate Gene Transcription. Molecular Oncology 6 (2012): 611-619.
- Morgan MA, Shilatifard A. Chromatin Signatures of Cancer. Genes and Development 29 (2015): 238-249.
- Harshman SW, Hoover ME, Huang C, et al. Histone H1 Phosphorylation in Breast Cancer. Journal of Proteome Research 13 (2014): 2453-2467.
- Thiagalingam SA, Cheng KH, Lee HJ, et al. Histone Deacetylases: Unique Players in Shaping the Epigenetic Histone Code. Annals of the New York Academy of Sciences 983 (2003): 84-100.
- Santos-Rosa H, Caldas C. Chromatin Modifier Enzymes, the Histone Code and Cancer. European Journal of Cancer 41 (2005): 2381-2402.
- Esteller M. Epigenetic changes in cancer. F1000 Biology Reports 3 (2011): 9.
- Hassler MR, Egger G. Epigenomics of Cancer-Emerging New Concepts. Biochimie 94 (2012): 2219-2230.
- Biswas S, Rao CM. Epigenetics in Cancer: Fundamentals and Beyond. Pharmacology and Therapeutics 173 (2017): 118-134.
- Nebbioso A, Tambaro FP, Dell’Aversana C, et al. Cancer Epigenetics: Moving Forward. PLoS Genetics 14 (2018): e1007362.
- Esteller Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Reviews Genetics 8 (2007): 286-298.
- Stirzaker C, Taberlay PC, Statham AL, et al. Mining Cancer Methylomes: Prospects and Challenges. Trends in Genetics 30 (2014): 75-84.
- Witcher M, Emerson BM. Epigenetic Silencing of the p16INK4a Tumor Suppressor is Associated with Loss of CTCF Binding and a Chromatin Boundary. Molecular Cell 34 (2009): 271-284.
- Funaki S, Nakamura T, Nakatani T, et al. Global DNA Hypomethylation Coupled to Cellular Transformation and Metastatic Ability. FEBS Letters 589 (2015): 4053-4060.
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The Human Colon Cancer Methylome Shows Similar Hypo-and Hypermethylation at Conserved Tissue-specific CpG Island Shores. Nature Genetics 41 (2009): 178-186.
- Khurana E, Fu Y, Chakravarty D, et al. Role of Non-coding Sequence Variants in Cancer. Nature Reviews Genetics 17 (2016): 93-108.
- Lin CP, He L. Noncoding RNAs in Cancer Development. Annual Review of Cancer Biology 1 (2017): 163-184.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA Networks in Cancer. Nature Reviews Cancer 18 (2018): 5-18.
- Ferreira HJ, Esteller M. Non-coding RNAs, Epigenetics, and Cancer: Tying It All Together. Cancer and Metastasis Reviews 37 (2018): 55-73.
- Formosa A, Lena AM, Markert EK, et al. DNA Methylation Silences miR-132 in Prostate Cancer. Oncogene 32 (2013): 127-134.
- He XX, Kuang SZ, Liao JZ, et al. The Regulation of microRNA Expression by DNA Methylation in Hepatocellular Carcinoma. Molecular BioSystems 11 (2015): 532-539.
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, A Non-protein-coding RNA, Controls Apoptosis and is Downregulated in Breast Cancer. Oncogene 28 (2009): 195-208.
- Hajjari M, Salavaty A. HOTAIR: An Oncogenic Long Non-coding RNA in Different Cancers. Cancer Biology and Medicine 12 (2015): 1-9.
- Bhan A, Mandal SS. LncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochimica et Biophysica Acta 1856(2015): 151-164.
- Qiu MT, Hu JW, Yin R, et al. Long Noncoding RNA: An Emerging Paradigm of Cancer Research. Tumor Biology 34 (2013): 613-620.
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Research 77 (2017): 3965-3981.
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. In Eds.: Allis CD, Jenuwein T, Reinberg D, et al. Epigenetics. Cold Spring Harbor, NY, Cold Spring Harbor Press (2007): 457-476.
- Allis CD, Jenuwein T. The Molecular Hallmarks of Epigenetic Control. Nature Reviews Genetics 17 (2016): 487-500.
- Flavahan WA, Gaskell E, Bernstein BE. Epigenetic Plasticity and the Hallmarks of Cancer. Science 357 (2017): 2380.
- Fouad YA, Aanei C. Revisiting the Hallmarks of Cancer. American Journal of Cancer Research 7 (2017): 1016-1036.
- Chatterjee A, Rodger EJ, Eccles MR. Epigenetic Drivers of Tumourigenesis and Cancer Metastasis. Seminars in Cancer Biology 51 (2018): 149-159.
- Burbee DG, Forgacs E, Zochbauer-Muller S, et al. Epigenetic Inactivation of RASSF1A in Lung and Breast Cancers and Malignant Phenotype Suppression. Journal of the National Cancer Institute 93 (2001): 691-699.
- Hannigan A, Smith P, Kalna G, et al. Epigenetic Downregulation of Human Disabled Homolog 2 Switches TGF-β from a Tumor Suppressor to a Tumor Promoter. Journal of Clinical Investigation 120 (2010): 2842-2857.
- Yanagawa N, Tamura G, Oizumi H, et al. Frequent Epigenetic Silencing of the p16 Gene in Non?small Cell Lung Cancers of Tobacco Smokers. Japanese Journal of Cancer Research 93 (2002): 1107-1113.
- Zheng J, Wang J, Sun X, et al. HIC1 Modulates Prostate Cancer Progression by Epigenetic Modification. Clinical Cancer Research 19 (2013): 1400-1410.
- Lyu T, Jia N, Wang J, et al. Expression and Epigenetic Regulation of Angiogenesis-related Factors During Dormancy and Recurrent Growth of Ovarian Carcinoma. Epigenetics 8 (2013): 1330-1346.
- Lujambio A, Esteller M. How Epigenetics can Explain Human Metastasis: A New Role for microRNAs. Cell cycle 8 (2009): 377-382.
- Langst G, Manelyte L. Chromatin Remodelers: From Function to Dysfunction. Genes 6 (2015): 299-324.
