Significant Aspect of Rv0378 Gene of Mycobacterium tuberculosis H37Rv Reveals the PE_PGRS like Properties by Computational Approaches
Article Information
Md Amjad Beg1,2, Fareeda Athar1, Laxman S Meena2*
1Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia, New Delhi, India
2CSIR-Institute of Genomics and Integrative Biology, Delhi, India
*Corresponding Author: Dr. Laxman S Meena, CSIR-Institute of Genomics and Integrative Biology Mall Road, Delhi-110007, India
Received: 18 February 2019; Accepted: 28 February 2019; Published: 06 March 2019
Citation:
Beg MA, Athar F, Meena LS. Significant Aspect of Rv0378 Gene of Mycobacterium tuberculosis H37Rv Reveals the PE_PGRS like Properties by Computational Approaches. Journal of Biotechnology and Biomedicine 2 (2019): 024-039.
Share at FacebookAbstract
Rv0378 is a conserved hypothetical gene of Mycobacterium tuberculosis H37Rv (M. tuberculosis H37Rv). This gene has been predicted to possess some PE_PGRS like characteristics. M. tuberculosis H37Rv contains a group of PE_PGRS qualities, with various identified motifs, encoding firmly related proteins that are outstandingly rich in glycine and alanine. These genes are predicted to be one of the key regulators for the pathogenic mechanism of this bacterium by the evidence of their absence in other a virulent strain. This manuscript noted some of the most valuable aspects about hypothetical protein Rv0378 being function as a PE_PGRS protein. The major insights of this study include interaction study, modelling by using I-TASSER, validation of the model by RAMPAGE analysis and prediction of metal binding sites by using Insilico approaches. The study shows that this gene is majorly a coiled structure with very short lengths and had a similarity with other already confirmed PE_PGRS proteins. The structure is validated by the bioinformatics approach. Interaction study by STRING shows that this protein interacts with its neighboring partner and secE2 protein which is a transporter protein. Prediction of ligand binding site confirms that this gene binds with calcium and magnesium with very high affinity which focus our step ahead in the direction of this gene as PE_PGRS protein and essential for pathogenesis. This study will be helpful in the experimental design to work in this field and if found correct as predicted, thus this work might be helpful in the direction of the treatment of this disease.
Keywords
Mycobacterium tuberculosis, Rv0378, PE_PGRS, Calcium (Ca2+), Macrophages
"Mycobacterium tuberculosis articles Mycobacterium tuberculosis Research articles Mycobacterium tuberculosis review articles Mycobacterium tuberculosis PubMed articles Mycobacterium tuberculosis PubMed Central articles Mycobacterium tuberculosis 2023 articles Mycobacterium tuberculosis 2024 articles Mycobacterium tuberculosis Scopus articles Mycobacterium tuberculosis impact factor journals Mycobacterium tuberculosis Scopus journals Mycobacterium tuberculosis PubMed journals Mycobacterium tuberculosis medical journals Mycobacterium tuberculosis free journals Mycobacterium tuberculosis best journals Mycobacterium tuberculosis top journals Mycobacterium tuberculosis free medical journals Mycobacterium tuberculosis famous journals Mycobacterium tuberculosis Google Scholar indexed journals " amino acid articles amino acid Research articles amino acid review articles amino acid PubMed articles amino acid PubMed Central articles amino acid 2023 articles amino acid 2024 articles amino acid Scopus articles amino acid impact factor journals amino acid Scopus journals amino acid PubMed journals amino acid medical journals amino acid free journals amino acid best journals amino acid top journals amino acid free medical journals amino acid famous journals amino acid Google Scholar indexed journals Biotechnology articles Biotechnology Research articles Biotechnology review articles Biotechnology PubMed articles Biotechnology PubMed Central articles Biotechnology 2023 articles Biotechnology 2024 articles Biotechnology Scopus articles Biotechnology impact factor journals Biotechnology Scopus journals Biotechnology PubMed journals Biotechnology medical journals Biotechnology free journals Biotechnology best journals Biotechnology top journals Biotechnology free medical journals Biotechnology famous journals Biotechnology Google Scholar indexed journals Rv0378 articles Rv0378 Research articles Rv0378 review articles Rv0378 PubMed articles Rv0378 PubMed Central articles Rv0378 2023 articles Rv0378 2024 articles Rv0378 Scopus articles Rv0378 impact factor journals Rv0378 Scopus journals Rv0378 PubMed journals Rv0378 medical journals Rv0378 free journals Rv0378 best journals Rv0378 top journals Rv0378 free medical journals Rv0378 famous journals Rv0378 Google Scholar indexed journals PE_PGRS articles PE_PGRS Research articles PE_PGRS review articles PE_PGRS PubMed articles PE_PGRS PubMed Central articles PE_PGRS 2023 articles PE_PGRS 2024 articles PE_PGRS Scopus articles PE_PGRS impact factor journals PE_PGRS Scopus journals PE_PGRS PubMed journals PE_PGRS medical journals PE_PGRS free journals PE_PGRS best journals PE_PGRS top journals PE_PGRS free medical journals PE_PGRS famous journals PE_PGRS Google Scholar indexed journals Calcium (Ca2+) articles Calcium (Ca2+) Research articles Calcium (Ca2+) review articles Calcium (Ca2+) PubMed articles Calcium (Ca2+) PubMed Central articles Calcium (Ca2+) 2023 articles Calcium (Ca2+) 2024 articles Calcium (Ca2+) Scopus articles Calcium (Ca2+) impact factor journals Calcium (Ca2+) Scopus journals Calcium (Ca2+) PubMed journals Calcium (Ca2+) medical journals Calcium (Ca2+) free journals Calcium (Ca2+) best journals Calcium (Ca2+) top journals Calcium (Ca2+) free medical journals Calcium (Ca2+) famous journals Calcium (Ca2+) Google Scholar indexed journals Macrophages articles Macrophages Research articles Macrophages review articles Macrophages PubMed articles Macrophages PubMed Central articles Macrophages 2023 articles Macrophages 2024 articles Macrophages Scopus articles Macrophages impact factor journals Macrophages Scopus journals Macrophages PubMed journals Macrophages medical journals Macrophages free journals Macrophages best journals Macrophages top journals Macrophages free medical journals Macrophages famous journals Macrophages Google Scholar indexed journals Fibronectin-binding articles Fibronectin-binding Research articles Fibronectin-binding review articles Fibronectin-binding PubMed articles Fibronectin-binding PubMed Central articles Fibronectin-binding 2023 articles Fibronectin-binding 2024 articles Fibronectin-binding Scopus articles Fibronectin-binding impact factor journals Fibronectin-binding Scopus journals Fibronectin-binding PubMed journals Fibronectin-binding medical journals Fibronectin-binding free journals Fibronectin-binding best journals Fibronectin-binding top journals Fibronectin-binding free medical journals Fibronectin-binding famous journals Fibronectin-binding Google Scholar indexed journals calcium-binding articles calcium-binding Research articles calcium-binding review articles calcium-binding PubMed articles calcium-binding PubMed Central articles calcium-binding 2023 articles calcium-binding 2024 articles calcium-binding Scopus articles calcium-binding impact factor journals calcium-binding Scopus journals calcium-binding PubMed journals calcium-binding medical journals calcium-binding free journals calcium-binding best journals calcium-binding top journals calcium-binding free medical journals calcium-binding famous journals calcium-binding Google Scholar indexed journals metabolism articles metabolism Research articles metabolism review articles metabolism PubMed articles metabolism PubMed Central articles metabolism 2023 articles metabolism 2024 articles metabolism Scopus articles metabolism impact factor journals metabolism Scopus journals metabolism PubMed journals metabolism medical journals metabolism free journals metabolism best journals metabolism top journals metabolism free medical journals metabolism famous journals metabolism Google Scholar indexed journals
Article Details
Abbreviations:
TB-Tuberculosis; M. tuberculosis H37Rv-Mycobacterium tuberculosis H37Rv; AMs-Alveolar macrophages; MDR-TB-Multidrug-Resistant TB; XDR-TB-Extremely Drug Resistant TB; PDB-Protein Data Bank; NCBI-National Center for Biotechnology Information; AA-amino acid; kD-kilo Dalton; bp-Base pair; Ca2+-Calcium
1. Introduction
Tuberculosis (TB) is a fatal disease which is caused by Mycobacterium tuberculosis H37Rv (M. tuberculosis H37Rv), which is aerobic and gram-positive bacteria [1]. Mycobacteria genus comprises of pathogenic and non-pathogenic strain like M. tuberculosis H37Rv, Mycobacterium bovis (M. bovis), Mycobacterium leprae (M. leprae), are pathogenic and non-pathogenic strain is Mycobacterium smegmatis (M. smegmatis). This pathogenic strain is available in the environment after coughing and sneezing of an already infected person and enters in the host through nasal track via inward breath as these pathogens are available in air droplets [2]. M. tuberculosis H37Rv is a facultative intracellular pathogen of macrophages. In case of TB this bacterium reaches to lungs where it endures for a longer time period in alveolar macrophages and successfully reside inside it [3]. In macrophages, it takes a longer period of time in the latent phase. M. tuberculosis H37Rv replicates in macrophages and get arranged with other immune cells which results in formation of granuloma like structure which is the hallmark of this disease [4]. In 2016, there are 6 million new cases showing resistance to Rifampicin [5], the most important valuable first-line drug to which 4.9 million had multidrug-resistant TB [MDR-TB] also known as the nastiest condition. Almost half of the cases of MDR-TB [47%] were in India, China and the Russian countries [6]. According to the 2017 WHO report, this report has developed a TB-Sustainable Development Goals to seek the outline of 14 indicators with the purpose of associated TB prevalence, under seven SDGs are the worldwide number of new and regress TB cases [7, 8]. Due to the persistence research and awareness curriculum, there will be 35% reduction in TB deaths and a 20% reduction in TB incidence in 2017, compared with levels in 2015. In overall world, South Asia, India and East Africa are devastated by TB [9-11]. For the ending of TB disease the first milestones of the End TB plan to be set for 2020. To understand the pathogenesis mechanism, the important thing is to describe the exacting highlights of M. tuberculosis H37Rv that make it powerful to avoid the host barrier framework and insert to its destructiveness [12].
In the M. tuberculosis H37Rv genome, there are multiple immunomodulatory proteins, including several members of PE_PPE family wherever PE_PGRS proteins are a subset. In M. tuberculosis H37Rv, there are 63 individual PE_PGRS family proteins are present and they are high GC rich content with repetitive homologues sequences [13-15]. PE_PGRS family proteins play a vital role in pathogenesis and there are three main domains, i.e. an N-terminal PE domain, repetitive PGRS domain and the unique C-terminal domain and it contributes to the protein localization on the mycobacterial surface [16]. Past few years ago, there are extensive work has been done to comprehend the part of PE_PGRS family proteins which is responsible in invasion, adhesion and cell surface marker of the host (macrophage and dendritic cells) [17]. In the early studies of PE_PGRS, we know that the proline-glutamic acid (PE) domain is responsible for the cellular localization of these proteins on bacterial cells [18]. Fibronectin-binding and calcium-binding property may be strongly implicated in immune-pathogenesis of virulent M. tuberculosis H37Rv strain [19, 20]. PE_PGRS proteins are variable surface antigens by the suggestion of some data analysis [21]. Previous studies had been proved the importance of PE_PGRS Family protein in complete virulence of this bacterium for example PE_PGRS 30 (Rv1651c) is necessary for the full virulence of M. tuberculosis H37Rv and enough for the stimulate cell death in host cells [22]. Here, we have explored the characteristics of the hypothetical glycine-rich protein of M. tuberculosis H37Rv Rv0378. PE_PGRS has glycine-rich sequence motif GGXGXD/NXUX, a nonapeptide sequence which predicts to the binding of Calcium (Ca2+) but still, the significance of these motifs is still unclear [23]. Rv0378 is a conserved hypothetical protein of this bacterium and contains pentapeptide (GGXGG) and Glycine content 41%. In Rv0378 protein, the PE_PGRS (GGXGG), calcium-binding motif (GGXGXD/NXUX) and guanine base recognition site (GSA) are present in its sequences as shown in Table 1.
|
S.No. |
Motifs |
Consensus Sequence |
Sequence |
Inter-Motif space (a.a) |
Reference |
|
1 |
PE-PGRS |
GGXGG |
23 GGAGG 27 |
- |
[15] |
|
29 GGAGG 33 |
2 |
||||
|
38 GGAGG 42 |
5 |
||||
|
2 |
Calcium Binding |
GGXGXD/NXUX |
23 GGAGGDGGS 31 |
- |
[24] |
|
38 GGEGGDAGA 46 |
7 |
||||
|
3 |
Buttreness the guanine base recognition site |
(T/G)(C/S)A |
15 GSA17 |
- |
[25] |
Table 1: The PE-PGRS, Calcium-binding motifs and Guanine base recognition site are identified in Rv0378 protein and their alignments.
The Ca2+ which also act like secondary messenger responsible for affecting the major cellular processes in eukaryotes by binding to proteins with varied affinities in both extracellular and intracellular environments [24, 25]. On the basis of previous reports, this bacterium has the ability to maintaining the Ca2+ homeostasis activity and it’s important for the prokaryotic physiology [26]. M. tuberculosis H37Rv included the capability to modify Ca2+ level in macrophages to favour their own metabolism [27]. In the phagosomes maturation and acidification, Ca2+ signalling are observed and their acidifications are restored by cytosolic Ca2+ via Ca2+ ionophore [28]. Therefore in this manuscript, we have used various Insilico approaches (which shown in supplementary Table S1) for evaluating the important perspective of Rv0378 protein of M. tuberculosis H37Rv which might empower experimental work in this field and thus found to be a suitable step in the development of antituberculosis drug [29, 30].
2. Methodology
2.1 Retrieval of the sequence Rv0378 protein
There are various databases which provided the genome sequence of M. tuberculosis H37Rv like KEGG, UniProt, NCBI, and Mycobrowser etc. For the retrieval of the Rv0378 gene, we use the UniProt (http://www.uniprot.org) where we retrieve the FASTA format sequence and physiochemical properties [31, 32].
2.2 Multiple sequence alignment
For the analysis of multiple sequence alignment was done by Clustal Omega. The latest tool of the Clustal family is CLUSTAL O (1.2.4). CLUSTAL O increases the considerable scalability more than previous versions, it allowing the large dataset to be aligned in merely a hardly some hours. CLUSTAL O have multiple processors, the quality of alignments is superior to earlier versions, as measured by a range of well-liked benchmarks [33]. The multiple sequence alignment of Rv0378 of M. tuberculosis H37Rv had been done with PE_PGRS61, PE_PGRS33, PE_PGRS10 and PE_PGRS3.
2.3 Secondary structure prediction
The secondary structure was predicted by using PSIPRED v3.3 (http://bioinf.cs.ucl.ac.uk) servers [34, 35].
2.4 Molecular modelling
Molecular modeling is a collection of methods for driving, manipulating and representing the structural demonstration of a biomolecule. The protein modeling of Rv0378 is done by the I-TASSER Iterative Threading ASSEmbly Refinement (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) server which is online available for predicting the homology modelling of the protein. I-TASSER modeller required the FASTA format of the protein sequence [36, 37]. I-TASSER server has done the protein modelling by following the three-stage while, the Local Meta threading server (LOMETS) for improving the secondary structure of the model protein by furtively after this secondary structure modelled they introduced the percentage or presence of alpha-helix, beta-sheet and coil region in the protein respectively. I-TASSER server, consider the modelled structure by the C-score, Z-score and coverage of threading alignment. The cutoff value is -5 to 2 for C-score, for the Z-score there are Z-score is greater than 1 and the Cov means the template coverage area of an alignment which is more than 70%. Statistically, a TM-score ≤ 0.17 corresponds to a similarity between two randomly selected structures from the PDB library; a TM-score >0.5 which means a similarity of the structure topology just approx similar [38, 39]. In this protein modeling, we have been doing the homology modelling and cross evaluation for the RaptorX (http://raptorx.uchicago.edu) and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2). For RaptorX there is the P-value, uGDT and GDT of the query arrangement with the best-positioned layout are exhibited to be utilized as evaluation of the quality of the subsequent model structure. GDT is figured as uGDT separated by the area length and duplicated by a 100. Here, uGDT is figured between the indigenous structure and the model built from the best-positioned layout. The cutoff value of the RaptorX displayed protein is p-value larger than 4, 95% of the models have uGDT more prominent than 50. Then again, if for estimations of p-value under 4, 98% of the models have uGDT under 50. This demonstrates if a model has uGDT larger than 50, which is acceptable [40, 41].
2.5 Validation of the modelled protein Rv0378
The evaluation of the modelled protein Rv0378 is validated by SAVES metaserver which have RAMPAGE, ERRAT and Verify3D for the refinement of modelled protein Rv0378. In SAVES server the RAMPAGE (http://services.mbi.ucla.edu/SAVES/Ramachandran/), assessed for its spine compliance utilizing a Ramachandran plot. RAMPAGE analyze the stereochemical quality of a protein structure by analyzing residue-by-residue geometry and overall structure geometry demonstrated the protein validate score for the validated result in most favoured region, additionally allowed region, generously allowed region and disallowed region [42, 43]. Verify3D (http://services.mbi.ucla.edu/Verify_3D/) was utilized to approve the refined structure. The 3D structure of the protein was contrasted with its own amino acid sequence thinking about a 3D profile ascertained from the nuclear directions of the structures of correct proteins [44] and the general characteristics of the demonstrated structures were assessed by the ERRAT (http://services.mbi.ucla.edu/ERRAT/) server [45]. A user can use all these programs in an input PDB structure or it can also do it individually run on the program one after one by the SAVES metaserver.
2.6 Binding site prediction
The anticipation of the active pocket of our showed protein Rv0378 is predictable by COACH online server. Prior to dynamic site-particular molecular docking (binding study), the confirmation of real pocket is crucial. Ligand can bound reversibly or irreversibly with the binding pocket site of Rv0378 protein. Only several amino deposits are in prosecuting with the ligand. Anyway, the other amino acids build up the protein is given precise arrangement and confirmation for the ligand binding site approach for COACH (https://zhanglab.ccmb.med.umich.edu/COACH/) server [46]. COACH is a meta-server, it starts from given structure of target protein then it will make correlative ligand binding site forecast using the two relative strategies, TM-site and S-site which perceive ligand-restricting course of action from the database (BioLiP) protein work database by restricting specific substructure and gathering profile examination [47].
2.7 Sub-cellular Localization Prediction
Protein localization studies are beneficiary for analysis, we could easily analyze by the TBpred server (http://crdd.osdd.net/raghava/tbpred/) which is an SVM based server to predict the Mycobacterial protein localization. TBpred predicted the protein localization is in the cytoplasm, integral membrane, lipid-anchored and secreted proteins. At last this server prediction accuracy module is 86.62% respectively [48].
2.8 Protein-protein interaction studies
To predict the interaction between protein-protein and chemical-protein is done by the STRING and STICH database. The interactions between protein-protein acquaintances as direct (physical) or indirect (functional) connections; STRING [49, 50] and STITCH [51, 52] could do the computational prediction; on the basis of sequence connection between the organisms, and from connections aggregated from other principal databases. STRING and STICH database interaction result shown separately for each data and the cutoff value as low confidence: scores <0.4; medium: 0.4 to 0.7; high: >0.7.
3. Result
3.1 Sequence retrieval
For the homology modelling of Rv0378 first, we know the FASTA format sequence which is retrieved from the UniProtKB (O53713_MYCTU) of the Rv0378. Rv0378 gene has 222 bp and ~6kD molecular mass of the protein. Rv0378 is conserved hypothetical glycine-rich protein which has PE_PGRS and Ca2+ binding motif which show PE_PGRS like properties.
3.2 Multiple sequence alignment
The sequence of Rv0378 gene of M. tuberculosis H37Rv shows the structural homology with PE_PGRS61, PE_PGRS33, PE_PGRS10 and PE_PGRS3 family protein. It was aligned with the PE_PGRS family protein of M. tuberculosis H37Rv by using CLUSTAL O (1.2.4) as shown in Figure 1.

Figure 1: Multiple sequence alignment.
Comparison of the Rv0378 gene of M. tuberculosis H37Rv with PE_PGRS61, PE_PGRS33, PE_PGRS10, and PE_PGRS3 of M. tuberculosis H37Rv. Identical amino acids are indicated by asterisks and the high similarity is indicated by a colon. A single dot denotes lesser similarities. Hyphens indicate gaps introduced to optimize alignment. The alignment was performed using the CLUSTAL O (1.2.4) Program. PE_PGRS pentapeptide is shown in bold and also marked by underline.
3.3 Secondary structure prediction
The secondary structure prediction analysis is done by PSIPRED shows the graphical representation of the percentage of the α-helices, β-sheets and coil region. PSIPRED server predicted to the secondary structure prediction of the protein analysis by the single amino acid sequence or FASTA format submission of the protein sequence. In Rv0378 protein, PSIPRED server predicted this protein is completely accommodating the coiled region which is clearly shown in Figure 2.
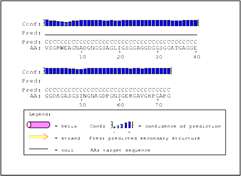
Figure 2: Secondary structure prediction of Rv0378 by PSIPRED tool.
The graphical output of PSIPRED prediction of secondary structure of the protein shows this protein have Coiled region 1-73 amino acid.
3.4 Molecular modelling
As we probably know for the protein modelling, first we retrieved the sequence for the UniProtKB ID (O53713) of Rv0378 protein which has 73 amino acid sequences. The Rv0378 protein appears as an approximate protein and had not previously studied. The Rv0378 protein modelling is based on homology modelling and it's done by I-TASSER, RaptorX and Phyre2 where the result is satisfactory. The homology modelling is done by I-TASSER threading the template by the MUSTER, FFAS-3D, SPARKS-X, HHSEARCH2, HHSEARCH I, Neff-PPAS, HHSEARCH, pGen THREADER, wdPPAS and PROSPECT2 threading programs. The threading template result shows Rv0378 protein template PDB hit is 3BOI and 2PNE whose identity is 44% coverage area of the template is 100% and the normalized z-score are 6.28 for 3BOI and 0.97 for 2PNE which is satisfactory and it’s shown in Table 2 and the top ten ranks identified structural analogues PDB of the Rv0378 is in Supplementary Table S2.
|
Rank |
PDB Hit |
Iden1 |
Iden 2 |
Cov |
Norm. Z-score |
|
1 |
3BOI |
0.32 |
0.44 |
1.00 |
6.28 |
|
2 |
2PNE |
0.34 |
0.44 |
1.00 |
0.97 |
|
3 |
2PNE |
0.35 |
0.44 |
0.99 |
6.87 |
|
4 |
3BOG |
0.48 |
0.45 |
0.97 |
1.74 |
|
5 |
3HR2 |
0.37 |
0.51 |
1.00 |
3.09 |
|
6 |
3BOI |
0.32 |
0.44 |
0.99 |
6.34 |
|
7 |
5CTD |
0.28 |
0.23 |
0.44 |
2.05 |
|
8 |
4DMU |
0.35 |
0.34 |
0.99 |
1.29 |
|
9 |
3BOI |
0.34 |
0.44 |
1.00 |
2.62 |
|
10 |
2PNE |
0.34 |
0.44 |
1.00 |
1.16 |
Table 2: Threading templates used by I-TASSER.
In model build for Rv0378 protein sequence we found top ten threading templates by I-TASSER server. The C-score is the certainty score for the model, which is from -5 to 2 where Rv0378 is -0.27 and the estimated TM-score, estimated RMSD is 0.68 ± 0.12 and 3.9 ± 2.6Å which is good enough. To find out the consensus prediction of the gene ontology of the Rv0378 protein which is done by the I-TASSER server showing molecular function, Biological process and Cellular component. The prediction of the gene ontology shows in Table 3.
The gene ontology prediction clearly shows this protein molecular function is carboxylic ester hydrolase activity with 42% accuracy and the biological process shows it is involved in the Metallo-sulfur cluster. The other homology tool RaptorX web server predicted the best template 2PNE which is also known earlier discussion in I-TASSER. The p-value of the RaptorX model structure is 8.41e-04 the overall uGDT is 56. RaptorX modelled the Rv0378 are 100% and the secondary structure is totally coiled all the 73 residues are predicted as a coil region. The result prediction by the Phyre2, template 3BOG and 2PNE confidence is 99.2 and 99 and this template is the antifreeze protein from organism Hypogastrura harveyi (H. harveyi). Finally, the displayed structure of the Rv0378 protein appears in Figure 3.
|
S.No. |
Molecular Function |
Biological Process |
Cellular Component |
|||
|
GO term |
GO score |
GO term |
GO score |
GO term |
GO score |
|
|
1 |
Carboxylic ester hydrolase activity |
0.42 |
The incorporation of a metal and exogenous sulfur into a metallo-sulfur cluster |
0.47 |
extracellular region |
0.67 |
|
2 |
Hydrolase activity, hydrolyzing O-glycosyl compounds |
0.40 |
A process that results in the biosynthesis and arrangement of constituent parts, or disassembly of a cell wall. |
0.47 |
- |
- |
|
3 |
- |
- |
The chemical reactions and pathways resulting in the breakdown of glucans, polysaccharides consisting only of glucose residues |
0.42 |
- |
- |
Table 3: Consensus prediction of GO terms.
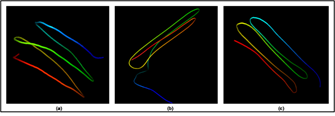
Figure 3: 3D protein modelling of Rv0378 via I-TASSER.
In this figure, subfigures (a-c) show the modelled protein Rv0378 3D structure construct via I-TASSER, RaptorX and Phyre2. (a) I-TASSER server determined by the C score, Estimated RMSD and Estimated TM-score. The determined score value of Rv0378 shows, C score is ?0.27, Estimated RMSD is 0.68 ± 0.12Å and Estimated TM-score is 3.9 ± 2.6 Å that prove RMSD of the I-TASSER models correlates better with C-score. (b) RaptorX modelled structure evaluated by the p-value of the model and uGDT. The modelled structure by RaptorX, the p-value is 8.41e-04 and the overall uGDT is 56. (c) At last, the Phyre2 server outcome template is 3BOG and 2PNE confidence is 99.2 and 99.
3.5 Evaluation of Rv0378 protein model
The protein model assessment by SAVES metaserver some server like (RAMPAGE, Verify3D and ERRAT). RAMPAGE (Ramachandran plot examination) showed that the most favoured region, additionally allowed region, generously allowed regions and residues in disallowed regions. These parameters of protein structure exhibiting that our showed protein was of good quality steady and acceptable. RAMPAGE analyzes factor is shown in Figure 4.

Figure 4: 3D model validation of Rv0378 protein.
In this figure, subfigures (a-c) showing the Ramachandran plot validation of the model by RAMPAGE. In the model protein validation (a) Modelled by I-TASSER server (b) by RaptorX and (c) Phyre2 showing Ramachandran plot analysis in most favoured region residues, residues in an additional allowed region, residues in a generously allowed region and residues in disallowed region. At last, for protein modelled by the RaptorX, validated result is most favoured region 94.9%, in the additional allowed region 2.6%, in the generously allowed region 2.6% in the disallowed region 0.0%.
The 3D profile evaluation of Rv0378 protein analysis is done by utilizing a Verify3D server. This program assesses the closeness of an atomic model (3D) with its own specific amino acid arrangement which is one dimensional. Each deposit is doled out an essential class in the brilliance of its zone and condition (alpha, beta, circle, polar, non-polar et cetera). At least 80% of the amino acids have scored >= 0.2 in the 3D/1D profile. 100.00% of the arrangement had found in the centre estimation of 3D-1D score >= 0.2 that is discerning for our exhibited protein appeared. ERRAT is an online server which favors the protein structure on the beginning of the atomic association between different sorts of molecules. The quality factor of Rv0378 protein structure is considered. The overall protein model assessment which is done by SAVES metaserver are shown in Table 4.
3.6 Structure-based binding site prediction of Rv0378
The prediction of the ligand binding site of the Rv0378 protein and it is done by the COACH server. They predicted probable ligand binding site which produces the consequent binding site forecasts utilizing the two similar strategies, TM-SITE and S-SITE. In ligand binding site prediction of Rv0378 protein, there are Calcium (Ca), Magnesium (Mg) and tetraethylene glycol monooctyl ether (C8E) are bound with protein as a ligand. By the estimation of COACH server, the results demonstrate on rank 1 Protein Data Bank (PDB) hit 4K1C which has C-score 0.08 where the cluster size is 4 and the Ligand name is Ca and the consensus binding residues are 6 and 46. In TM-SITE and S-SITE result rank 1, the PDB ID is 3SY9 but the C-score 0.18 and cluster size 2 and the ligand is C8E where the predicted binding sites are 30, 31 and 45. At last, the server demonstrate S-SITE result where the c-score 0.23, cluster size 6 their ligand is Ca and Mg and the predicted binding sites are 2, 4, 6, 8, 9, 10, 11, 12, 13, 14, 15, 21, 22, 23, 26, 33, 34, 39, 40, 41, 42, 44, 49, 52, 53, 54, 57 they are all the ligand residue sites are shown in Figure 5.
|
S.No. |
Model |
RAMPAGE |
Verify 3D |
ERRAT |
|
1 |
I-TASSER |
Residues in most favoured region 53.8% Residues in additional allowed region 30.8%Residues in generously allowed region 10.3%Residues in disallowed region 5.1% |
100.00% (PASS) |
78.125 |
|
2 |
RaptorX |
Residues in most favoured region 94.9% Residues in additional allowed region 2.6%Residues in generously allowed region 2.6%Residues in disallowed region 0.0% |
100.00% (PASS) |
0 |
|
3 |
PHYRE2 |
Residues in most favoured region 89.7% Residues in additional allowed region 7.7%Residues in generously allowed region 0.0%Residues in disallowed region 2.6% |
92.40% (PASS) |
23.076 |
Table 4: Model evaluation by SAVES (Statistical Analysis and Verification Server).
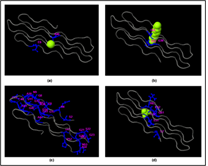
Figure 5: Ligand binding sites prediction by COACH server.
Rv0378 protein-ligand binding pocket predicted by Coach server shows the Calcium (CA), Tetraethylene glycol monooctyl ether (C8E) Magnesium (MG) and Phosphate (PO4) binding ligand in and these subfigure (a) this result shows CA ligand predicted consensus binding residue are 6 and 46; (b) C8E ligand predict by TM-SITE and the binding position is 30, 31 and (c) In S-SITE result shows the predicted ligand CA and MG and their binding sites are 2, 4, 6, 8, 9, 10, 11, 12, 13, 14, 15, 21, 22, 23, 26, 33, 34, 39, 40, 41, 42, 44, 49, 52, 53, 54, 57 and (d) At last the Cofactor outcome showing the ligand PO4 predicted binding residue are 2, 31, 32, 61, 62 and 63.
3.7 Localization prediction
According to TBpred server, selected on the basis of Dipeptide composition based SVM, the Rv0378 protein is attached to the membrane by lipid anchor with the satisfactory score positive value 0.10340302 as shown in Figure 6.
Figure 6: Subcellular localization prediction.
TBpred server predicted the localization of Rv0378 protein; it's predicted as attached to the membrane by a lipid anchor.
3.8 Protein-protein interaction studies
The prediction of the protein-protein and protein chemical interaction by the STRING database and STITCH database server which results shows the Rv0378 gene interacts with the secE2 and Rv0377 genes. secE2 (Rv0379) which are M. tuberculosis H37Rv species and transport protein it binds with calcium ions and might play role in sequestering additional small ligand and Rv0377 is LysR family transcriptional regulator. They both gene are neighborhoods interaction. The scoring value of STRING and STITCH database-secE2 is 0.566 and Rv0377 is 0.468 by STRING and secE2 is 0.737 and Rv0377 is 0.611 which shown in Figure 7.
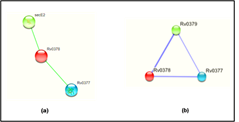
Figure 7: Interaction of the modelled protein Rv0378 by STRING & STITCH tool.
STRING and STITCH server predicts the interacting partner of the protein with their competence of the interaction between them. This figure shows (a) the protein-protein interaction of Rv0378 by STRING which result are secE2 is 0.566 and Rv0377 is 0.468 and (b) the protein-chemical interaction by STITCH which also shows the same interacting partner but score high value as secE2 is 0.737 and Rv0377 is 0.61 with cutoff value of this server is [0-1].
4. Discussion
In the present study with respect to these discoveries leads us to think about the threat of enhancement of the illness in a new populace. The hazard is on ascending because of inaccessibility of a compelling medication. In spite of the actuality that the number of occurrences had been diminished the vulnerability builds step by step equally because of a few natural factors. There is constant exertion had been put by researchers with the end goal to build the adequacy of the immunization and in looking for a new approach to find a drug target. In this manuscript, we accentuate the characteristic and modeling of the Rv0378 protein of M. tuberculosis H37Rv and various bioinformatics approaches have been applied to this gene to intricate its different function and essentiality in a biological system. Rv0378 protein is about 6 kDa and a glycine-rich sequence which is retrieved from UniProtKB [31, 32], comprising calcium binding motif, PE_PGRS motif and guanine base recognition site which predicts that Rv0378 protein has PE_PGRS like properties. For the multiple sequence alignment of Rv0378 result shows the alignment with PE_PGRS61, PE_PGRS33, PE_PGRS10 and PE_PGRS3 family protein [33]. The secondary structure prediction by the PSIPRED showed this 2D structure of protein is coiled region, all the 73 residues are coil loop [34, 35]. The 3D structure modelled by I-TASSER, RaptorX and Phyre2 which have a 3BOI and 2PNE template which have coverage of the template is 100 % identity of the template is 44% and the normalized z-score is 6.28 this study finding the various root of the protein. Rv0378 modelled protein describe in an earlier study this 3D model validated by the SAVES metaserver which shows most favoured region 94.9%, in the additional allowed region 2.6%, in the generously allowed region 2.6% in the disallowed region 0.0% [37-39].
For the ligand binding site predicted by the COACH server which result shows the ligand are Ca, Mg and PO4 and the residue where the ligand bind 6, 30, 31, 45, 61, 62 and 63 [46, 47]. This protein is localized in the membrane where it is attached with lipid anchor [48]. Rv0378 interact with the secE2 (a transport protein) which binds with calcium ions, might play role in sequestering additional small ligand and Rv0377 (LysR family transcriptional regulator), both genes are neighborhoods interaction [49-52]. After analysis of Rv0378, we find that this protein might be a role as a PE_PGRS like properties that seem to be essential for the vitality of M. tuberculosis H37Rv inside the phagosomes of the host [22]. As we discussed earlier PE_PGRS is one of the important class of proteins, understanding and classification of these genes might be an important step in knowing the strategy of the survival of this bacterium inside the host cell and therefore could be used a step in the eradication of this disease [53]. Additionally, investigation of this study may turn out with the cohort of successful medication treatment against this deadly disease.
5. Conclusion
The current situation of tuberculosis (TB) level, we wanted a secure way to prevent our generation vulnerability situation. This manuscript highlighting the characteristics of Rv0378 protein of M. tuberculosis H37Rv features PE_PGRS like properties. PE_PGRS proteins are one of the key regulators for virulence of this bacterium and are very much important class to study. Featuring characteristics might be helpful in the experimental study about this gene. This gene also found to be linked with Ca and Mg metals and this interaction shows its significance towards the pathogenesis of this bacterium. Further bioinformatics studies might help in understanding the mechanistic action of the Rv0378 protein work which might be approachable output as an affirmative result towards the treatment of this disease.
Acknowledgement
The authors acknowledge financial support from the Department of Science and Technology-SERB, Council of Scientific and Industrial Research-Institute of Genomics and Integrative Biology under the research project GAP0145. Md. Amjad Beg also acknowledges University Grants Commission Maulana Azad National Fellowship for the financial support and Jamia Millia Islamia University.
Conflict of Interest
There is no conflict of interest.
References
- Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nature Reviews Molecular Cell Biology 2 (2001): 569-577.
- Barry CE, Boshoff HI, Dartois V, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nature Reviews Molecular Cell Biology 7 (2009): 845-855.
- Esmail H, Barry CE, Young DB, et al. The ongoing challenge of latent tuberculosis. Philos Trans R Soc Lond B Biol Sci 369 (2014): 20130437.
- Pieters J. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host and Microbe 3 (2008): 399-407.
- World Health Organization. Global Tuberculosis Report 2015 (2016).
- Cohen KA, Bishai WR, Pym AS. Molecular basis of drug resistance in Mycobacterium tuberculosis. Microbiology Spectrum 2 (2014).
- World Health Organization. Global Tuberculosis Report 2015 (2017).
- Pai M, Behr MA, Dowdy D, et al. Tuberculosis. Nature Reviews Disease Primers 2 (2016).
- Duarte TA, Nery JS, Boechat N, et al. A systematic review of east african-indian family of Mycobacterium tuberculosis in brazil. Brazilian Journal of Infectious Diseases 3 (2017): 317-324.
- Aliannejad R, Bahrmand A, Abtahi H, et al. Accuracy of a new rapid antigen detection test for pulmonary tuberculosis. Iranian Journal of Microbiology 8 (2016): 238-242.
- Yerlikaya S, Broger T, MacLean E, et al. A Tuberculosis Biomarker Database: The Key to Novel TB Diagnostics. International Journal of Infectious Diseases 56 (2017): 253-257.
- Telenti A, Imboden P, Marchesi F, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341 (1993): 647-650.
- Meena LS, Meena J. Cloning and characterization of a novel PE_PGRS60 protein (Rv3652) of Mycobacterium tuberculosis H37Rv exhibit fibronectin-binding property. Biotechnology and Applied Biochemistry 63 (2016): 525-531.
- Monu, Meena LS. Biochemical characterization of PE_PGRS61 family protein of Mycobacterium tuberculosis H37Rv reveals the binding ability to fibronectin. Iranian Journal Basic Medical Science 19 (2016): 1105-1113.
- Flores J, Espitia C. Differential expression of PE and PE_PGRS genes in Mycobacterium tuberculosis Gene 318 (2003): 75-81.
- De Maio F, Maulucci G, Minerva M, et al. Impact of protein domains on PE_PGRS30 polar localization in Mycobacteria. PLoS One 9 (2014): e112482.
- Meena LS. An overview to understand the role of PE_PGRS family proteins in Mycobacterium tuberculosis H37Rv and their potential as new drug targets. Biotechnology and Applied Biochemistry 62 (2015): 145-53.
- Cascioferro A, Delogu G, Colone M, et al. PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Molecular Microbiology 66 (2007): 1536-1547.
- Meena PR, Monu, Meena LS. Fibronectin binding protein and Ca2+ play an access key role to mediate pathogenesis in Mycobacterium tuberculosis: An overview. Biotechnology and Applied Biochemistry 63 (2016): 820-826.
- Espitia C, Laclette JP, Mondragón-Palomino M, et al. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology 145 (1999): 3487-3495.
- Banu S, Honore N, Saint-Joanis B, et al. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Molecular Microbiology 44 (2002): 9-19.
- Iantomasi R, Sali M, Cascioferro A, et al. PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis. Cellular Microbiology 14 (2012): 356-367.
- Yeruva VC, Kulkarni A, Khandelwal R, et al. The PE_PGRS Proteins of Mycobacterium tuberculosis Are Ca(2+) Binding Mediators of Host-Pathogen Interaction. Biochemistry 55 (2016): 4675-4687.
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 294 (2001): 1299-1304.
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nature Reviews Molecular Cell Biology 1 (2000): 11-21.
- Knight MR, Campbell AK, Smith SM, et al. Recombinant aequorin as a probe for cytosolic free Ca2+ in Escherichia coli. FEBS Letters 282 (1991): 405-408.
- Vergne I, Chua J, Singh SB, et al. Cell biology of Mycobacterium tuberculosis Annual Review of Cell and Developmental Biology 20 (2004): 367-394.
- Malik ZA, Denning GM, Kusner DJ. Inhibition of Ca(2+) signaling by Mycobacterium tuberculosis is associated with reduced phagosome-lysosome fusion and increased survival within human macrophages. The Journal of Experimental Medicine 191 (2000): 287-302.
- Beg M A, Shivangi, Thakur SC, et al. Structural prediction and mutational analysis of Rv3906c gene of Mycobacterium tuberculosis H37Rv to determine its essentiality in survival. Advances in Bioinformatics 2018 (2018): 6152014.
- Shivangi, Beg A, Meena S, et al. To Find out the Essentiality of Rv0526 Gene in Virulence of Mycobacterium Tuberculosis by using In silico Approaches. Open Journal of Bacteriology 1 (2017): 013-015.
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Research 45 (2017): 158-169.
- UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Research 43 (2015): 204-212.
- Sievers F, Wilm A, Dineen D, et al. Fast scalable generationof high quality protein multiple sequence alignment using Clustal Omega. Molecular Systems Biology 7 (2011): 539.
- Monu, Meena LS. Biochemical characterization of PE_PGRS61 family protein of Mycobacterium tuberculosis H37Rv reveals the binding ability to fibronectin. Iranian Journal of Basic Medical Science 19 (2016): 1105-1113.
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 16 (2000): 404-405.
- Jones DT. Protein structure prediction in the postgenomic era. Current Opinion of Structure Biology 10 (2000): 371-379
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9 (2008): 40.
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein and function prediction. Nature Protocols 5 (2010): 725-38.
- Yang J, Yan R, Roy A, et al. The I-TASSER Suite: protein structure and function prediction prediction. Nature Methods 12 (2015): 7-8.
- Ma J, Wang S, Zhao F, et al. Protein threading using context-specific alignment potential. Bioinformatics 29 (2013): 257-265.
- Peng J, Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins 79 (2011): 161-71.
- Lovell SC, Davis IW, Arendall WB, et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Protiens 50 (2003): 437-450.
- Ho BK, Brasseur R. The Ramachandran plots of glycine and pre-proline. BMC Structure Biology 5 (2005): 14.
- Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Science 2 (1993): 1511-1519.
- Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253 (1991):164-170.
- Wang S, Sun S, Li Z, et al. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Computational Biology 13 (2017): e1005324.
- Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29 (2013): 2588-2595.
- Rashid M, Saha S, Raghava GP. Support Vector Machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinformatics 8 (2007): 337.
- Von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Research 31 (2003): 258-261.
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research 45 (2017): 362-368.
- Kuhn M, Szklarczyk D, Pletscher-Frankild S, et al. STITCH 4: integration of protein-chemical interactions with user data. Nucleic Acids Research 42 (2014): 401-407.
- Szklarczyk D, Santos A, von Mering C, et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research 44 (2016): 380-384.
- Rajni, Meena LS. Survival mechanisms of pathogenic Mycobacterium tuberculosis FEBS Journal 277 (2010): 2416-2427.
