SETD2 Deficiency and Mir-21: Potent Therapeutic Targets in NUT Midline Carcinoma
Article Information
Nana Yoshida1, Shunsuke Okumura1*, Takaaki Sasaki1, Shin-Ichi Chiba2, Masatoshi Sado3, Kyohei Oyama4, Ryohei Yoshida1, Noriko Hirai1, Yoshinori Minami1, Masahiro Kitada1, Yoshinobu Ohsaki1
1Respiratory Center, Asahikawa Medical University-Asahikawa, Hokkaido, Japan
2Center for Adanced Researh and Education, Asahikawa Medical University-Asahikawa, Hokkaido, Japan
3Surgical Pathology, Asahikawa Medical University Hospital-Asahikawa, Hokkaido, Japan
4Division of Cardiovascular Surgery, Asahikawa Medical University-Asahikawa, Hokkaido, Japan
*Corresponding Author: Shunsuke Okumura, Asahikawa Medical University, 2-1-1-1 Midorigaoka-Higashi, Asahikawa, Hokkaido 0788510, Japan.
Received: 28 February 2022; Accepted: 07 March 2022; Published:> 14 March 2022
Citation:
Nana Yoshida, Shunsuke Okumura, Takaaki Sasaki, Shin-Ichi Chiba, Masatoshi Sado, Kyohei Oyama, Ryohei Yoshida, Noriko Hirai, Yoshinori Minami, Masahiro Kitada, Yoshinobu Ohsaki. SETD2 deficiency and miR-21: Potent therapeutic targets in NUT midline carcinoma. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 106-120.
Share at FacebookAbstract
Nuclear Protein in Testis (NUT) Midline Carcinoma (NMC) is a rare and highly aggressive tumor with the bromodomain containing 4 proteins (BRD4)-NUT (NUTM1) gene fusion. Few targeted therapies are available for NMC, and thus novel therapeutic targets are required. To the best of our knowledge, the present study was the first to report that SET domain-containing protein 2 (SETD2) deficiency and microRNA (miRNA/miR)-21 may serve as novel therapeutic targets for the treatment of NMC. First, Next-Generation Sequencing (NGS) identified a novel SETD2 mutation (p.Ser2382fs) in the NMC cell lines, HCC2429 and Ty82. Trimethylation of lysine 36 on histone H3 expression was depleted in the NMC cells, which was indicative of SETD2 loss. NMC cells were sensitive to the WEE1 G2 checkpoint kinase (WEE1) inhibitor, AZD1775, in the cancer cells with SETD2 deficiency. NMC cells that were resistant to Bromodomain and Extra-Terminal Motif (BET) inhibitors were next established, and these resistant cells were also sensitive to AZD1775. Subsequently, miRNA analysis revealed that miR-21 expression was increased in BET-inhibitor-resistant NMC cells. In addition, miR-21 regulated the proliferation of NMCs. The miR-21 inhibitor also suppressed the proliferation of BET-inhibitor-resistant cells. Additionally, a digital PCR assay was established to detect NUT gene rearrangements to identify patients with NMC. A total of 32 clinical samples were analyzed and one case of NMC was identified, in which the novel SETD2 mutation was detected. These data suggested that SETD2 loss and miR-21 may be therapeutic targets for treatment of NMC.
Keywords
MicroR-21; Nuclear protein in testis midline carcinoma, SET domain-containing protein 2, WEE1 G2 checkpoint kinase
MicroR-21 articles; Nuclear protein in testis midline carcinoma articles, SET domain-containing protein 2 articles, WEE1 G2 checkpoint kinase articles
MicroR-21 articles MicroR-21 Research articles MicroR-21 review articles MicroR-21 PubMed articles MicroR-21 PubMed Central articles MicroR-21 2023 articles MicroR-21 2024 articles MicroR-21 Scopus articles MicroR-21 impact factor journals MicroR-21 Scopus journals MicroR-21 PubMed journals MicroR-21 medical journals MicroR-21 free journals MicroR-21 best journals MicroR-21 top journals MicroR-21 free medical journals MicroR-21 famous journals MicroR-21 Google Scholar indexed journals Nuclear protein in testis midline carcinoma articles Nuclear protein in testis midline carcinoma Research articles Nuclear protein in testis midline carcinoma review articles Nuclear protein in testis midline carcinoma PubMed articles Nuclear protein in testis midline carcinoma PubMed Central articles Nuclear protein in testis midline carcinoma 2023 articles Nuclear protein in testis midline carcinoma 2024 articles Nuclear protein in testis midline carcinoma Scopus articles Nuclear protein in testis midline carcinoma impact factor journals Nuclear protein in testis midline carcinoma Scopus journals Nuclear protein in testis midline carcinoma PubMed journals Nuclear protein in testis midline carcinoma medical journals Nuclear protein in testis midline carcinoma free journals Nuclear protein in testis midline carcinoma best journals Nuclear protein in testis midline carcinoma top journals Nuclear protein in testis midline carcinoma free medical journals Nuclear protein in testis midline carcinoma famous journals Nuclear protein in testis midline carcinoma Google Scholar indexed journals SET domain-containing protein 2 articles SET domain-containing protein 2 Research articles SET domain-containing protein 2 review articles SET domain-containing protein 2 PubMed articles SET domain-containing protein 2 PubMed Central articles SET domain-containing protein 2 2023 articles SET domain-containing protein 2 2024 articles SET domain-containing protein 2 Scopus articles SET domain-containing protein 2 impact factor journals SET domain-containing protein 2 Scopus journals SET domain-containing protein 2 PubMed journals SET domain-containing protein 2 medical journals SET domain-containing protein 2 free journals SET domain-containing protein 2 best journals SET domain-containing protein 2 top journals SET domain-containing protein 2 free medical journals SET domain-containing protein 2 famous journals SET domain-containing protein 2 Google Scholar indexed journals WEE1 G2 checkpoint kinase articles WEE1 G2 checkpoint kinase Research articles WEE1 G2 checkpoint kinase review articles WEE1 G2 checkpoint kinase PubMed articles WEE1 G2 checkpoint kinase PubMed Central articles WEE1 G2 checkpoint kinase 2023 articles WEE1 G2 checkpoint kinase 2024 articles WEE1 G2 checkpoint kinase Scopus articles WEE1 G2 checkpoint kinase impact factor journals WEE1 G2 checkpoint kinase Scopus journals WEE1 G2 checkpoint kinase PubMed journals WEE1 G2 checkpoint kinase medical journals WEE1 G2 checkpoint kinase free journals WEE1 G2 checkpoint kinase best journals WEE1 G2 checkpoint kinase top journals WEE1 G2 checkpoint kinase free medical journals WEE1 G2 checkpoint kinase famous journals WEE1 G2 checkpoint kinase Google Scholar indexed journals aggressive tumor articles aggressive tumor Research articles aggressive tumor review articles aggressive tumor PubMed articles aggressive tumor PubMed Central articles aggressive tumor 2023 articles aggressive tumor 2024 articles aggressive tumor Scopus articles aggressive tumor impact factor journals aggressive tumor Scopus journals aggressive tumor PubMed journals aggressive tumor medical journals aggressive tumor free journals aggressive tumor best journals aggressive tumor top journals aggressive tumor free medical journals aggressive tumor famous journals aggressive tumor Google Scholar indexed journals bromodomain articles bromodomain Research articles bromodomain review articles bromodomain PubMed articles bromodomain PubMed Central articles bromodomain 2023 articles bromodomain 2024 articles bromodomain Scopus articles bromodomain impact factor journals bromodomain Scopus journals bromodomain PubMed journals bromodomain medical journals bromodomain free journals bromodomain best journals bromodomain top journals bromodomain free medical journals bromodomain famous journals bromodomain Google Scholar indexed journals novel therapeutic targets articles novel therapeutic targets Research articles novel therapeutic targets review articles novel therapeutic targets PubMed articles novel therapeutic targets PubMed Central articles novel therapeutic targets 2023 articles novel therapeutic targets 2024 articles novel therapeutic targets Scopus articles novel therapeutic targets impact factor journals novel therapeutic targets Scopus journals novel therapeutic targets PubMed journals novel therapeutic targets medical journals novel therapeutic targets free journals novel therapeutic targets best journals novel therapeutic targets top journals novel therapeutic targets free medical journals novel therapeutic targets famous journals novel therapeutic targets Google Scholar indexed journals microRNA articles microRNA Research articles microRNA review articles microRNA PubMed articles microRNA PubMed Central articles microRNA 2023 articles microRNA 2024 articles microRNA Scopus articles microRNA impact factor journals microRNA Scopus journals microRNA PubMed journals microRNA medical journals microRNA free journals microRNA best journals microRNA top journals microRNA free medical journals microRNA famous journals microRNA Google Scholar indexed journals Next-Generation Sequencing articles Next-Generation Sequencing Research articles Next-Generation Sequencing review articles Next-Generation Sequencing PubMed articles Next-Generation Sequencing PubMed Central articles Next-Generation Sequencing 2023 articles Next-Generation Sequencing 2024 articles Next-Generation Sequencing Scopus articles Next-Generation Sequencing impact factor journals Next-Generation Sequencing Scopus journals Next-Generation Sequencing PubMed journals Next-Generation Sequencing medical journals Next-Generation Sequencing free journals Next-Generation Sequencing best journals Next-Generation Sequencing top journals Next-Generation Sequencing free medical journals Next-Generation Sequencing famous journals Next-Generation Sequencing Google Scholar indexed journals Trimethylation articles Trimethylation Research articles Trimethylation review articles Trimethylation PubMed articles Trimethylation PubMed Central articles Trimethylation 2023 articles Trimethylation 2024 articles Trimethylation Scopus articles Trimethylation impact factor journals Trimethylation Scopus journals Trimethylation PubMed journals Trimethylation medical journals Trimethylation free journals Trimethylation best journals Trimethylation top journals Trimethylation free medical journals Trimethylation famous journals Trimethylation Google Scholar indexed journals
Article Details
1. Introduction
Nuclear Protein in Testis (NUT) Midline Carcinoma (NMC), also referred to as NUT carcinoma, is a rare and highly aggressive tumor that predominantly affects midline structures, which occurs in both children and adults. NMC is genetically identified by the presence of the NUT gene, also known as the NUT midline carcinoma family member 1 (NUTM1) gene rearrangement [1, 2]. The most frequent translocation in NMC is observed between the NUT gene at chromosome 15q14 and the Bromodomain Containing Protein 4 (BRD4) gene on chromosome 19q13.1 chromosome, which accounts for >70% of the rearrangements [3-5]; other NUT fusion partners include BRD3, Nuclear Receptor Binding SET Domain Protein 3 (NSD3) and zinc finger protein 532 (ZNF532), amongst other genes [6-9]. NMC lacks specific pathological features and can occur in any organ, such as the thorax, head and neck, or other midline organs [10]. Patients with NMC can be misdiagnosed with other malignant tumors due to the poor differentiation, and lack of awareness of the disease symptoms and diagnostic tests available. The prognosis of patients with NMC is considerably poor, with a median survival time of 6–9 months [3, 10]. The development of novel treatment strategies for NMC is challenging. Targeted therapy has been performed based on the molecular mechanisms underlying aberrant signaling through the fusion proteins. BRD4 is a member of the bromodomain and Extra-Terminal Domain (BET) family of proteins and binds to acetylated lysine in histones [11]. BET inhibitors have been investigated, and the first-in-class BET inhibitor, JQ1, which competitively binds to bromodomains, has shown prodifferentiative and antiproliferative effects on NMC in preclinical studies [12]. More recently, novel BET inhibitors have been developed and studied in clinical trials [13, 14]. However, the efficacy of these inhibitors are limited, and NMC may develop resistance to them [14, 15]. To date, no other therapeutic targets have been identified in NMC. Accordingly, the development of novel treatment strategies is required. Recently, increased onco-microRNA (miRNA/miR) expression has been reported in NMC [16]. Studies that focus on the presence of simultaneous gene mutations and miRNAs may assist in identifying novel therapeutic targets. In the present study, the role of two epigenetic regulators, SET Domain Containing 2 (SETD2) and miR-21, as possible therapeutic targets for NMC, were investigated.
2. Material and Methods
2.1. Cell culture and reagents
NMC cell lines, Ty82 (RIKEN Cell Bank), HCC2429 cells (UT Southwestern Medical Center) and A549 lung adenocarcinoma cells (American Type Culture Collection) were incubated in RPMI-1640 medium supplemented with 10% FBS and antibiotics at 37°C with 5% CO2 [17, 18]. BET inhibitors, JQ-1 and OTX015 (MK8628; Birabresib) and the WEE1 G2 checkpoint kinase (WEE1) inhibitor, AZD1775 (MK1775; Adavosertib) were purchased from Selleck Chemicals. The BET-inhibitor-resistant HCC2429 cell line was established by exposure to increasing concentrations of JQ-1 (1-100 nM) over a 3-months period, as described previously [19].
2.2. MTS assay
Cell proliferation assays were performed using the CellTiter 96 Aqueous One Solution assay (Promega Corporation). Briefly, cells were seeded into 96-well plates (3-5x103 cells/well) overnight in triplicates. Following the incubation, the cells were treated with the indicated concentrations of drugs for 3 days, then the assays were performed. The experiments were performed independently at least three times.
2.3. Transfection
Stealth RNAi Small Interfering (si)RNA (Thermo Fischer Scientific, Inc.) was used for NUTM1 knockdown, Specifically, NUTM1-siRNAs (cat. no. HSS138007, HSS138008 and HSS138009) and Stealth RNAi siRNA Negative Control (cat. no. 12935300, Thermo Fischer Scientific, Inc.) were used. Lipofectamine® RNAiMAX transfection reagent and Opti-MEM I reduced serum medium (Thermo Fischer Scientific, Inc.) were used for siRNA transfection. The cells were treated with 50 pmol of siRNA for the indicated time periods, and subsequent experiments were then performed. miRNA function analysis was performed using miRCURY LNA miRNA mimics and inhibitors (all from Qiagen, Inc.); specifically hsa-miR-21-5p inhibitor (cat. no. YI04100689), hsa-miR-21-5p mimic (cat. no. YM00473093), miRNA inhibitor controls, and miRNA mimic controls. Lipofectamine® 3000 (Thermo Fischer Scientific, Inc.) and Opti-MEM I reduced serum medium were used for miRNA transfection. Cells were plated into 96-well plates and treated with 0.66 pmol of miRNA mimics or 5 pmol of miRNA inhibitors for 72 h, and subsequent assays were performed.
2.4 Western blotting
NUT (cat. no. 3625), b-actin (cat. no. 4967), and horseradish peroxidase (HRP)-labeled anti-rabbit IgG (cat. no. 7074) antibodies were purchased from Cell Signaling Technology, Inc. Trimethylation of lysine 36 on histone H3 (H3K36me3; cat. no. ab9050) and Histone H3 (cat. no. ab1791) antibodies were purchased from Abcam. Proteins were extracted from the cells using cell lysis buffer (Csll Signalint Technology, Inc.) supplemented with complete protease inhibitor cocktail (Roche Diagnostics). All antibodies were used at a dilution of 1:1,000. NuPAGE gels, Pierce Power Blotter, and iBind Western system (Thermo Fischer Scientific, Inc.) were used for western blotting, according to the manufacturers’ protocols. Protein expression was measured using the WSE-6100 LuminoGraph I (ATTO Corporation) and quantified using the CS Analyzer version 4 (ATTO Corporation).
2.5. Next-generation sequencing
Genomic DNA was extracted from the cell lines using the Blood & Cell Culture DNA mini kit (Qiagen, Inc.) and from patient-derived tumors using the QIAamp DNA FFPE Tissue kit (Qiagen, Inc.). DNA was quantified using the Qubit 2.0 fluorometer with the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Inc.). A total of 50 ng of DNA from cell lines and 10 ng of DNA from tissue samples were used for PCR amplification using the Ion AmpliSeq Library kit 2.0, and Ion AmpliSeq Comprehensive Cancer Panel (Thermo Fischer Scientific, Inc.). The Ion Express Barcode Adapters (Thermo Fischer Scientific, Inc.) were ligated to the PCR products for barcoding the tissue samples. AMPure XP beads (Beckman Coulter, Inc.) were used for PCR product purification. The libraries were sequenced using an Ion PGM system (Thermo Fischer Scientific, Inc.). DNA sequencing data were obtained using the Torrent Suite version 4.0 software (Thermo Fischer Scientific, Inc.). Variant caller version 4.0. was used to call variants. The reads were aligned with the GRCh38 reference genome, and Ion Reporter version 5.0 and CLC Genomics Workbench version 9.5.1 (Qiagen, Inc.) were used for additional analysis. Small RNAs were extracted from the NMC cell lines using a PureLink miRNA isolation kit (Thermo Fischer Scientific, Inc.) and quantified using the Agilent 2100 Bioanalyzer with an Agilent RNA small RNA kit (Agilent Technologies, Inc.). A small RNA library was prepared using the Ion Total RNA-Seq kit v2 (Thermo Fischer Scientific, Inc.), and the libraries were sequenced on the Ion PGM system. The reads were aligned with miRBase miRNAs. miRNA expression was analyzed using the CLC Genomics Workbench version 9.5.1.
2.6. miRNA assay
Small RNAs were extracted from NMC cells using the PureLink miRNA Isolation kit and quantified using a NanoDrop spectrophotometer (Thermo Fischer Scientific, Inc.). miRNA expression was analyzed using TaqMan microRNA assays (Thermo Fischer Scientific, Inc.), mir-21-5p (cat. no. 000397) and RNU48 (cat. no. 001006). In the present study, RNU48 was used as a housekeeping miRNA. Reverse Transcription (RT) was performed using the TaqMan microRNA Reverse Transcription kit (Thermo Fischer Scientific, Inc.) and the indicated RT primers in the assay kits. PCR was performed using the QuantStudio 3D Digital PCR Master Mix, QuantStudio 3D Digital PCR 20 K Chip kit v2, and ProFlex 2x Flat PCR system (Thermo Fisher Scientific, Inc.). PCR was performed according to the manufacturer’s instructions [20]. Absolute quantification was performed using the QuantStudio 3D Digital PCR system (Thermo Fischer Scientific, Inc.). We analyzed the data using the QuantStudio 3D AnalysisSuite Cloud Software (Thermo Fischer Scientific, Inc.).
2.7. Detection of the BRD4-NUT fusion genes in clinical samples
The Medical Ethics Committee of the Asahikawa Medical University approved the clinical sample analysis (approval no. 15218-2). All methods were performed in accordance with the relevant guidelines and regulations. Clinical samples [Formalin-Fixed Paraffin-Embedded (FFPE) tissues] from patients with a thoracic tumor were collected at the Asahikawa Medical University Hospital. Written informed consent was obtained from all patients before the tumor sample collection. The specific primers and probes for BRD4-NUT detection were designed and the sequences were: BRD4-NUT forward, 5'-TGAAGGGCTTCTCGTCCTCAG-3' and reverse, 5'-GCGGCACTAGGTTTCATGCTC-3'; and BRD4-NUT FAM probe, 5'-TCGGAGAGCTCAGTGAGTCCAGCT-3'. An RNeasy Mini kit (Qiagen, Inc.) was used to extract RNA from the cell lines and the RecoverAll Total Nucleic Acid Isolation kit for FFPE (Thermo Fischer Scientific, Inc.) was used to extract RNA from the clinical samples. First-strand cDNA was synthesized using the SuperScript VILO cDNA Synthesis kit (Thermo Fischer Scientific, Inc.), and the RT reaction was performed according to the manufacturer’s protocol. The QuantStudio 3D Digital PCR Master Mix, QuantStudio 3D Digital PCR 20 K Chip Kit v2, and ProFlex 2x Flat PCR System were used for PCR amplification according to the manufacturer’s protocol. Absolute quantification was performed using the QuantStudio 3D Digital PCR system, and QuantStudio 3D Analysis Suite Cloud software was used for data analysis [21].
2.8. Statistical Analysis
Data are presented as the mean ± SEM. Statistical analysis was performed using the GraphPad Prism Software version 7.0 (GraphPad Software, Inc.), and the results were analyzed using a Student’s t-test, One-way ANOVA, and Tukey’s multiple comparisons test. A Two-sided P<0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. SETD2 loss-of-function mutation is present in NMC
To date, concomitant mutations other than fusion genes have not been fully elucidated in NMC. Additionally, the fusion protein regulates the cell proliferation and epigenesis of NMC, and the role of epigenetic regulators, such as miRNA, remains to be studied in NMC. Therefore, gene mutations and miRNA expression was assessed using NGS to identify novel therapeutic targets for NMC. First, gene mutational analysis was performed using the NMC cell lines, HCC2429 and Ty82. Using a comprehensive cancer panel for NGS, 10 non-synonymous mutations were identified in HCC2429 cells, and 19 were identified in Ty82 cells (Figure 1a). Amongst these mutations, a focus was placed on the novel SETD2 mutation, SETD2-p.Ser2382fs, as SETD2 is an epigenetic regulator. Furthermore, this mutation was common to both cell lines. SETD2 is a tumor suppressor gene [22-24]. Physiologically, SETD2 is a non-redundant trimethyltransferase responsible for the trimethylation of lysine 36 on histone H3 (H3K36me3). The frameshift mutation was located on exon 17 before the WW and Set2 Rpb1 Interacting (SRI) domains of the SETD2 gene (Figure 1b). Hence, it was inferred that the mutation may cause the loss of SETD2 function; that is, decreased H3K36me3 levels in NMC. Indeed, western blotting analysis revealed decreased levels of H3K36me3 in NMC cells, particularly in HCC2429 cells (Figure 1c).
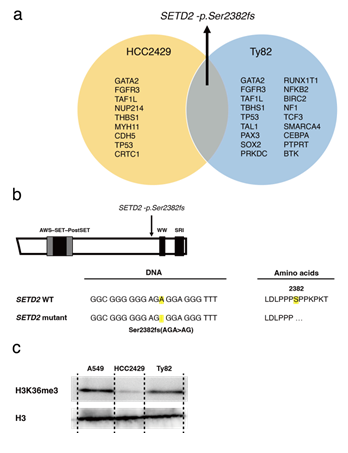
Figure 1: NMC cell lines with the recurrent SETD2 mutation. a. A list of concurrent gene mutations in the NMC cell lines HCC2429 and Ty82. Next-generation sequencing identified the recurrent SETD2-p.Ser2382fs mutation. b. The SETD2 frameshift mutation in the HCC2429 cells. The mutation is located before the WW domain. c. H3K36me3 levels as determined by western blotting. H3K36me3 levels were lower in NMC cells compared with the A549 cell line, a lung adenocarcinoma cell line. H3K36me3 and H3 levels were studied in A549, HCC2429 and Ty82 cell lines. Groups are separated by dividing lines. NMC, nuclear protein in testis midline carcinoma; H3K36me3, trimethylation of lysine 36 on histone H3; SETD2, SET domain-containing protein 2. There is synthetic lethality between H3K36me3 deficiency and WEE1 inhibition: H3K36me3-deficient cancers are significantly sensitive to WEE1 inhibitors, which induce DNA damage in tumors [25]. Next, the anti-tumor efficacy of the WEE1 inhibitor, AZD1775, in NMC cells was evaluated. The cell viability assay showed that NMC cells were sensitive to AZD1775 compared with A549 cells, which were used as a control lung cancer cell line harboring wild-type SETD2 (Figure 2a).
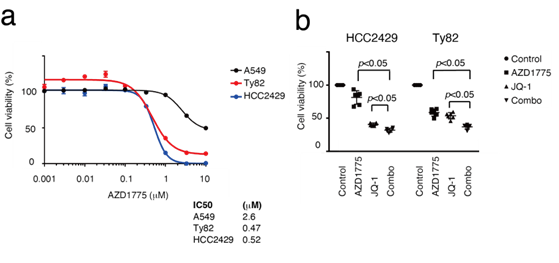
Figure 2: NMC cells are sensitive to the WEE1 inhibitor, AZD1775. a. MTS assays comparing sensitivity to AZD1775 between NMC and A549 cells. Cells were treated with the indicated concentrations of AZD1775 for 3 days. The NMC cells were sensitive to AZD1775 compared with A549. b. MTS assays studying the efficacy of the combination with AZD1775 and a BET inhibitor JQ-1. HCC2429 cells were treated with 0.3 mM AZD1775, 30 nM JQ-1 or both combined for 3 days. Ty82 cells were treated with 1.0 mM AZD1775, 30 nM JQ-1, or both combined for 3 days. BET, bromodomain and extra-terminal domain; WEE1, WEE1 G2 checkpoint kinase; NMC, nuclear protein in testis midline carcinoma.
The proliferation suppressive effects of the combination of AZD1775 and JQ-1 were next assessed (Figure 2b). As a result, additive effects of the combination were observed, suggesting that the efficacy of WEE1 inhibition is independent of BET inhibition. NMC often acquires resistance to BET inhibitors. Thus, HCC2429 cells resistant to BET inhibitors (HCC2429-JQR) were established by sustained treatment with JQ-1, and the HCC2429-JQR cells were found to be resistant to both JQ-1 and another BET inhibitor, OTX015 (MK8628, Birabresib) (Figure 3a). Cell proliferation assays showed that even HCC2429-JQR cells were sensitive to AZD1775 (Figure 3b).
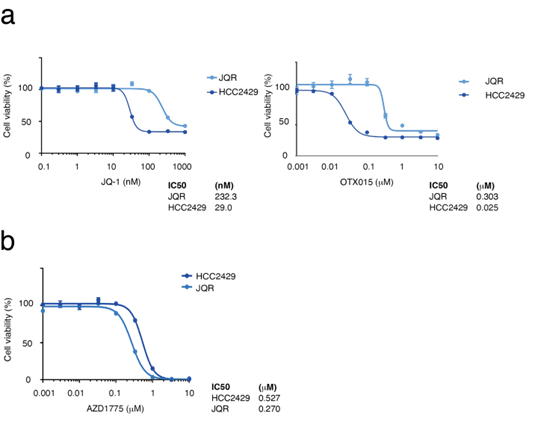
Figure 3: BET-inhibitor-resistant HCC2429 cells (HCC2429-JQR) are sensitive to AZD1775. a. MTS assays comparing sensitivity to BET inhibitors in the parent HCC2429 and HCC2429-JQR cells. Cells were treated with the BET inhibitors JQ-1 and OTX015 for 3 days. HCC2429-JQR cells were resistant to BET inhibitors. b. MTS assays in HCC2429 and HCC2429-JQR cells. These cells were treated with AZD1775 for 3 days. HCC2429-JQR cells were sensitive to AZD1775. BET, bromodomain and extra-terminal domain.
3.2. miR-21 regulates the proliferation of NMC
miRNA expression analyses were performed next. miRNAs are key regulators of epigenetics and pathogenesis in almost every type of cancer [26]. However, the relationship between miRNAs and BRD4-NUT has not been fully elucidated. To determine changes in miRNA expression using BRD4 or NUT inhibition, RNA-seq was performed on HCC2429 cells transfected with NUT siRNA or JQ-1. Using western blotting, it was determined that HCC2429 cells transfected with NUT siRNA exhibited decreased NUT expression (Figure 4a). The MTS assay showed that NUT siRNA had negligible effects on the proliferation of HCC2429 cells (Figure 4a). However, RNA-seq analysis revealed that the expression levels of miRNA were notably altered between the siRNA-transfected and untransfected cells; the most common miRNA was let-7a-1/let-7a-2/let-7a-3 in untransfected cells, whereas miR-21-5p (hereafter referred to as miR-21) was the most abundant miRNA in siRNA-transfected cells (Figure 4b). In contrast, JQ-1 did not significantly alter miR-21 expression in HC2429 cells (Figure 4b). miR-21 is an oncomiR, thus it was hypothesized that miR-21 may be associated with NMC cell proliferation [27]. The cell proliferation effects of miR-21 mimics or inhibitors on NMC were studied. miR-21 mimics increased the proliferation of HCC2429 cells; however, miR-21 inhibitors decreased their proliferation (Figure 4c). The transfection efficiency with miR-21 mimics and inhibitors was shown in supplementary (Figure 1, 2).
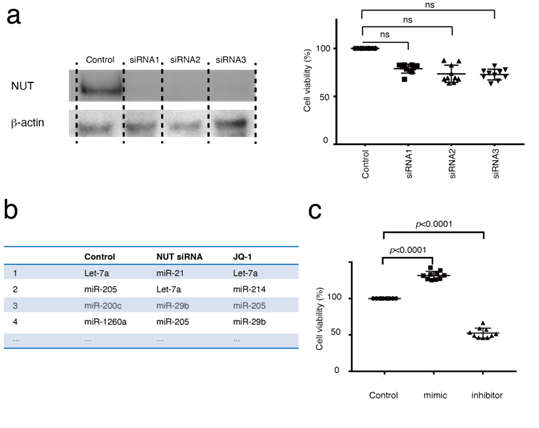
Figure 4: Altered miR-21 expression was observed in HCC2429, and miR-21 regulates HCC2429 cell proliferation. (a) Efficacy of NUT knockdown in HCC2429. HCC2429 cells were transfected with NUT siRNAs for 3 days. Western blotting demonstrated decreased NUT expression following NUT knockdown. NUT and b-actin levels were studied in HCC2429 cells following NUT knockdown. The groups are separated by dividing lines. MTS assays showed partial suppression of cell proliferation using NUT siRNAs. (B) miRNA expression profiles in HCC2429. RNA sequencing was performed in HCC2429 following NUT knockdown or following treatment with JQ-1. Compared with the control cells, miR-21 expression was increased in the NUT-siRNA-treated cells. (C) MTS assays studying the effects of miR-21 mimic or miR-21 inhibitor in HCC2429. The miR-21 mimic and inhibitor regulated the proliferation of HCC2429 cells. siRNA, small interfering RNA; miRNA/miR, microRNA; NUT, nuclear protein in testis.
Short-term treatment with JQ-1 did not alter the expression levels of miR-21 in NMCs (Figure 4b). However, it was hypothesized that miR-21 may be related to BET inhibitor resistance, as studies have shown that tumors with acquired resistance to BET inhibitors do not have gatekeeper mutations and drug pump activation [28]. Next, miR-21 expression in HCC2429-JQR cells was determined using the TaqMan miRNA assay. miR-21 expression was increased in the HCC2429-JQR cells compared with that in the parental cells (Figure 5a). An MTS assay demonstrated that the miR-21 inhibitor suppressed the proliferation of HCC2429-JQR cells (Figure 5b). Since AZD1775 suppressed cell proliferation in the HCC2429-JQR cells as described, the efficacy of the combination with the miR-21 inhibitor and AZD1775 was assessed. Notably, this combination exhibited additive effects on HCC2429-JQR cells (Figure 5c).
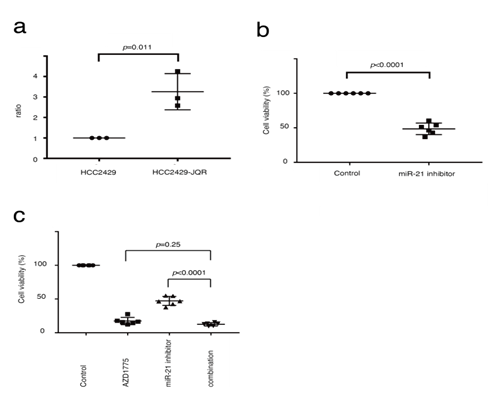
Figure 5: miR-21 expression is increased in HCC2429-JQR cells. (A). miRNA-based assays in HCC2429 and HCC2429-JQR cells. miR-21 expression was significantly increased in HCC2429-JQR cells compared with the parental HCC2429 cells. (B) MTS assays studying the efficacy of the miR-21 inhibitor in HCC2429-JQR cells. HCC2429-JQR cells were treated with the miR-21 inhibitor for 3 days. The miR-21 inhibitor suppressed the proliferation of HCC2429-JQR cells. (C) MTS assays in HCC2429-JQR cells treated with a combination of AZD1775 and miR-21 inhibitors. The HCC2429-JQR cells were treated with 1 mM AZD1775, the miR-21 inhibitor or both combined for 3 days. miRNA/miR, microRNA.
3.3. SETD2 loss-of-function mutations are present in a patient with NMC
Finally, gene mutations in clinical NMC samples were investigated. As the thorax is the most frequent location of NMC, the expression of the NUT fusion genes in patients with malignant thoracic tumors, whose tumors were located in the midline of their bodies were investigated [29]. A total of 32 tumor samples collected via transbronchial biopsy were retrospectively analyzed. Screening for NMC is performed using Immunohistochemistry (IHC), which finds that NUT is expressed in the nuclei of NMC cells [30, 31]. However, in some cases, IHC analysis cannot be performed due to the lack of tissue, especially in small biopsy samples. In the present study, most of the biopsy samples were very small, and it was assumed that these samples included a small number of cancer cells. A highly sensitive digital (d) PCR assay was established to detect the BRD4-NUT fusion gene. In the preclinical study, the dPCR assay successfully detected the BRD4-NUT fusion gene in HCC2429 cells; the dPCR assay detected a 1,000-fold diluted fusion gene (Figure 6a). As a result, one NMC tumor amongst the 32 tumors was detected using the assay (Figure 6b). As the recurrent SETD2 mutation was observed in the NMC cell lines, gene mutational analysis was performed to detect the SETD2 mutation in the NMC tumor, using the cancer comprehensive panel for NGS; and the SETD2-p.Ser2382fs mutation was detected in the tumor.
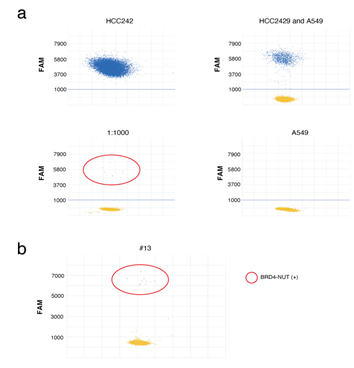
Figure 6: dPCR assays to identify BRD4-NUT in NMC. a. dPCR assays of HCC2429 and A549 cells. Specific probes for BRD4-NUT (FAM) were designed. The probe successfully identified BRD4-NUT in HCC2429, at up to a 1,000-fold dilution. b. dPCR assays of clinical samples. The assays identified the BRD4-NUT in a tumor. dPCR, digital PCR; NUT, nuclear protein in testis; NMC, NUT midline carcinoma; BRD4, bromodomain-containing protein 4.
4. Discussion
In the present study, two epigenetic regulators, SETD2 and miR-21, were identified as potential therapeutic targets for management of NMC, regardless of resistance to BET inhibitors. To the best of our knowledge, this is the first report showing the efficacy of in vitro targeted therapy with WEE1 and miR-21 inhibitors in NMC cells. First, a novel SETD2 mutation in NMC was identified. Somatic mutations of the SETD2 gene have been reported in several types of cancer, in which loss of SETD2 function leads to decreased H3K36me3 levels [22, 32, 33]. SETD2 has three conserved domains, AWS-SET-PostSET, WW, and Set2 Rpb1 Interacting (SRI) domains [34-36]. Taken together, SETD2 plays a key role in homologous recombination repair and genome stability by catalyzing trimethylation at H3K36 [37]. In the present study, the novel frameshift mutation was determined to be located just before the WW domain, suggesting loss of SETD2 functions as SRI domain deficiency abolishes trimethylation of H3K36me3 [38,39]. Indeed, H3K36me3 expression was decreased and AZD1775 was efficacious when used to treat HCC2429 cells, whereas H3K36me3 expression and effects of AZD1775 were less potent in Ty82 cells. There may be possible mechanisms that compensate for H3K36me3 in Ty82 cells, and further studies are required to assess this possibility. Overall, SETD2 loss may lead to genomic instability in NMC. Identification of the SETD2 mutation can lead to the development of targeted therapies for NMC. In H3K36me3-deficient tumors, WEE1 inhibition has a synthetic lethal interaction with H3K36me3 loss; the WEE1 inhibitor AZD1775 selectively kills SETD2-deficient cancer cells through dNTP starvation as a result of ribonucleotide reductase regulatory subunit M2 depletion [25]. In the present study, the NMC cells were more sensitive to AZD1775 than the BET inhibitors, which was consistent with the results of the present study. Next, it was shown that miR-21 regulated the proliferation of NMC cells. A previous study showed a set of 48 dysregulated miRNAs in NMC, in which the miRNAs targeting critical genes other than BRD4 and NUT were analyzed; however, miR-21 was not included in the 48 miRNAs [16]. Another study screened an miRNA mimic library and identified miR-3140, which was shown to target and suppress BRD4 by binding to its coding sequence [40]. Another report analyzed miRNA expression in NMC using clinical samples that identified three cases of sinonasal NMC, and two out of three NMCs showed upregulation of miR-21, miR-143, and miR-484 expression [41]. miRNA expression is regulated by DNA methylation and histone modifications [42]. Therefore, SETD2 deficiency may be associated with miR-21 expression.
The present study could not find a relationship between miR-21 and SETD2 using miRDB (mirdb.org/index.html), but altered promoter methylation of miR-21 has been reported in SETD2-deficient cancers [32]. In the present study, combination chemotherapy had additive beneficial effects even on BET-inhibitor-resistant NMC. A recent report showed that the combination therapy targeting BET and p300 was more effective than the BET inhibitor alone [43]. The combination of target inhibitors in another epigenetic category may be useful for the treatment of NMC. Taken together, AZD1775 and miR-21 inhibition may be used to overcome resistance to BET inhibitors in NMC. The present study has several limitations. Although it was demonstrated that AZD1775 and the miR-21 inhibitor suppressed the proliferation of NMC cells, it was not clarified whether they regulated in vitro cell migration and invasion. It remains to be determined whether the SETD2 mutation is recurrent in NMC. In addition, mutant SETD2 function and structure should be assessed. Finally, the present study did not perform in vivo experiments, which would enhance the credibility of the in vitro results. These considerations will be taken into account in future experiments.
5. Conclusion
In conclusion, the present study identified SETD2 deficiency and miR-21 as therapeutic targets for management of NMC. WEE1 and miR-21 inhibitors may serve as novel therapeutic options for the treatment of NMC.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grants-in-Aid for Scientific Research), #18K08164.
Competing Interests
The authors declare that they have no competing interests.
References
- French CA, Miyoshi I, Aster JC, et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am J Pathol 159 (2001): 1987-1992.
- French CA, Miyoshi I, Kubonishi I, et al. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res 63 (2003): 304-307.
- French CA. NUT Carcinoma: Clinicopathologic features, pathogenesis, and treatment. Pathol Int 68 (2018): 583-595.
- Bauer DE, Mitchell CM, Strait KM, et al. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res 18 (2012): 5773-5779.
- Chau NG, Hurwitz S, Mitchell CM, et al: Intensive treatment and survival outcomes in NUT midline carcinoma of the head and neck. Cancer 122 (2016): 3632-3640.
- French CA, Ramirez CL, Kolmakova J, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 27 (2008): 2237-2242.
- French CA, Rahman S, Walsh EM, et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov 4 (2014): 928-941.
- Alekseyenko AA, Walsh EM, Zee BM, et al. Ectopic protein interactions within BRD4-chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad Sci USA 114 (2017): 4184-4192.
- Shiota H, Elya JE, Alekseyenko AA, et al. Z4 Complex Member Fusions in NUT Carcinoma: Implications for a Novel Oncogenic Mechanism. Mol Cancer Res 16 (2018): 1826-1833.
- French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol 7 (2012): 247-265.
- Dhalluin C, Carlson JE, Zeng L, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399 (1999): 491-496.
- Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature 468 (2010): 1067-1073.
- Noel JK, Iwata K, Ooike S, et al. Abstract C244: Development of the BET bromodomain inhibitor OTX015. Molecular Cancer Therapeutics 12 (2013): 244-244.
- Lewin J, Soria JC, Stathis A, et al. Phase Ib Trial With Birabresib, a Small-Molecule Inhibitor of Bromodomain and Extraterminal Proteins, in Patients With Selected Advanced Solid Tumors. J Clin Oncol 36 (2018): 3007-3014.
- Piha-Paul SA, Hann CL, French CA, et al. Phase 1 Study of Molibresib (GSK525762), a Bromodomain and Extra-Terminal Domain Protein Inhibitor, in NUT Carcinoma and Other Solid Tumors. JNCI Cancer Spectr 4 (2020): 93.
- Pathak E, Bhavya, Mishra D, et al. Deciphering the Role of microRNAs in BRD4-NUT Fusion Gene Induced NUT Midline Carcinoma. Bioinformation 13 (2017): 209-213.
- Stirnweiss A, Oommen J, Kotecha RS, et al Molecular-genetic profiling and high-throughput. Oncotarget 8 (2017): 112313-112329.
- Wang R, Cao XJ, Kulej K, et al. Uncovering BRD4 hyperphosphorylation associated with cellular transformation in NUT midline carcinoma. Proc Natl Acad Sci USA 114 (2017): 5352-5361.
- Tsuji T, Ozasa H, Aoki W, et al. YAP1 mediates survival of ALK-rearranged lung cancer cells treated with alectinib via pro-apoptotic protein regulation. Nat Commun 11 (2020): 74.
- Conte D, Verri C, Borzi C, et al. Novel method to detect microRNAs using chip-based QuantStudio 3D digital PCR. BMC Genomics 16 (2015): 849.
- Santos RB, Pires AS, Abranches R. Addition of a histone deacetylase inhibitor increases recombinant protein expression in Medicago truncatula cell cultures. Sci Rep 7 (2017): 16756.
- Li J, Duns G, Westers H, et al SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget 7 (2016): 50719-50734.
- Fahey CC, Davis IJ. SETting the Stage for Cancer Development: SETD2 and the Consequences of Lost Methylation. Cold Spring Harb Perspect Med 7 (2017): 026468.
- McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci 74 (2017): 3317-3334.
- Pfister SX, Markkanen E, Jiang Y, et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell 28 (2015): 557-568.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16 (2017): 203-222.
- Bautista-Sánchez D, Arriaga-Canon C, Pedroza-Torres A, et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol Ther Nucleic Acids 20 (2020): 409-420.
- Shu S, Lin CY, He HH, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529 (2016): 413-417.
- French CA. The importance of diagnosing NUT midline carcinoma. Head Neck Pathol 7 (2013): 11-16.
- Haack H, Johnson LA, Fry CJ, et al. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol 33 (2009): 984-991.
- Park HS, Bae YS, Yoon SO, et al. Usefulness of Nuclear Protein in Testis (NUT) Immunohistochemistry in the Cytodiagnosis of NUT Midline Carcinoma: A Brief Case Report. Korean J Pathol 48(2014): 335-338.
- Network CGAR: Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499 (2013): 43-49.
- Chiang YC, Park IY, Terzo EA, et al. Haploinsufficiency for Microtubule Methylation Is an Early Driver of Genomic Instability in Renal Cell Carcinoma. Cancer Res 78 (2018): 3135-3146.
- Gao YG, Yang H, Zhao J, et al Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin. Structure 22 (2014): 378-386.
- Li M, Phatnani HP, Guan Z, et al. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA 102 (2005): 17636-17641.
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27 (2008): 406-420.
- Pfister SX, Ahrabi S, Zalmas LP, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 7 (2014): 2006-2018.
- Kizer KO, Phatnani HP, Shibata Y, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25 (2005): 3305-3316.
- Li B, Howe L, Anderson S, et al. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278 (2003): 8897-8903.
- Tonouchi E, Gen Y, Muramatsu T, et al. miR-3140 suppresses tumor cell growth by targeting BRD4 via its coding sequence and downregulates the BRD4-NUT fusion oncoprotein. Sci Rep 8 (2018): 4482.
- Laco J, Kovarikova H, Chmelarova M, et al. Analysis of DNA methylation and microRNA expression in NUT (nuclear protein in testis) midline carcinoma of the sinonasal tract: a clinicopathological, immunohistochemical and molecular genetic study. Neoplasma 65 (2018): 113-123.
- Handa H, Murakami Y, Ishihara R, et al. The Role and Function of microRNA in the Pathogenesis of Multiple Myeloma. Cancers (Basel) 11 (2019): 1738.
- Morrison-Smith CD, Knox TM, Filic I, et al. Combined Targeting of the BRD4-NUT-p300 Axis in NUT Midline Carcinoma by Dual Selective Bromodomain Inhibitor, NEO2734. Mol Cancer Ther 19 (2020): 1406-1414.
