Regulation of TGF-β1-Smad-1-7 signaling to Inhibit Epithelial Mesenchymal Transition by Repurposed Anti-Fibrotic Drug Pirfenidone can Attenuate Lung Cancer Progression
Article Information
Ritu Kulshrestha1*, Pawan Kumar1, Amit Singh1, Divya S Nair1, Meenu R1, Himanshu Dhanda1, Apoorva Pandey1, Dahiya R1, Mishra AK2, Dinda AK3
1Department of Pathology, V.P.Chest Institute, University of Delhi, India
2Institute of Nuclear Medicine & Allied Sciences, DRDO, New Delhi-110 054, Delhi, India
3Department of Pathology, All India Institute of Medical Sciences and Emeritus Professor, Indian Council of Medical Research, New Delhi-110 029, Delhi, India
*Corresponding Author: Ritu Kulshrestha, Department of Pathology, V.P.Chest Institute, University of Delhi, India.
Received: 16 September 2022; Accepted: 16 January 2023; Published: 28 February 2023
Citation: Ritu Kulshrestha, Pawan Kumar, Amit Singh, Divya S Nair, Meenu R, Himanshu Dhanda, Apoorva Pandey, Dahiya R, Mishra AK, Dinda AK. Regulation of TGF-β1-Smad-1-7 signaling to Inhibit Epithelial Mesenchymal Transition by Repurposed Anti-Fibrotic Drug Pirfenidone can Attenuate Lung Cancer Progression. Journal of Cancer Science and Clinical Therapeutics. 7 (2023): 50-64.
Share at FacebookAbstract
Background: Lung cancer cells undergo EMT, and metastasize to preferential sites such as brain, bone and adrenal glands. Pirfenidone is an anti-fibrotic agent used for treatment of pulmonary fibrosis. Its efficacy as therapeutic adjuvant in lung cancer via regulation of TGF-β1-Smad-1-7- EMT signaling is postulated.
Method: Lung adenocarcinoma (A-549) cells were cultured in DMEM at 37°C in 5% CO2. Cells were treated with saline (Control), bleomycin (20 mM) and Bleomycin+Pirfenidone (500μg/ml) and harvested at 4, 6, 8, 24, and 48h. The time-course of bleomycin induced TGF-β-BMPSmad- 1-7 signaling, EMT (E-Cadherin and Vimentin) and their inhibition by pirfenidone was assessed.
Results: Bleomycin caused a bimodal upregulation of TGF-β1 at 6, 48 h and EMT of A-549 cells (progressive E-cadherin downregulation, vimentin upregulation). An associated differential upregulation of (i) receptor Smads-(rSmads-2,3,5): Smad-2,5 (Bimodal upregulation at 4,48 hrs), Smad-3 (persistent upregulation- 4 to 48hrs), (ii) Co-Smad-4-persistent upregulation from 4 to 48 hrs, (iii) iSmad-1, 6,7: Smad-1 (upregulation at 24,48 hrs), Smad-6,7- early up-regulation at 4, 6 hrs respectively, was seen. Pirfenidone significantly reversed EMT by downregulating the TGF-β1/Smad-2-5 signaling, Vimentin expression and upregulating BMPiSmads- 6,7 signaling, E-Cadherin expression. This reduced cell migration.
Conclusion: Pirfenidone attenuates lung cancer cell migration by differentially inhibiting the TGF-β1-Smads-1-7 signaling pathway and EMT. Thus indicating the utility of pirfenidone as an adjuvant therapy to attenuate lung cancer progression and improve the prognosis of lung cancer patients on chemotherapy.
Keywords
A-549 Cell Line; Epithelial Mesenchymal Transition; TGF– β1-Smad-1-7 signaling
Article Details
1. Introduction
Lung cancer is the leading cause of cancer-related mortality in both men and women. Poor prognostic factors of lung cancer include advanced stage and presence of metastasis at the time of diagnosis [1]. Cancer progression is characterized by cell migration toward higher oxygen tension. This is characterised with changes in the: (i) cell structure (epithelial mesenchymal transition (EMT), (ii) loss of cell–cell adhesions and cell orientation, (iii) acquiring cancer stem-cell like properties with reduction of epithelial markers (cytokeratin-8, E-cadherin) and gain of mesenchymal phenotype (vimentin, α-SMA) (iii) altered interaction with tumor microenvironment (fibroblasts/myofibroblasts, endothelial cells), resulting in increased motility and invasion, (iv) altered tumor cell communication with the immune system (macrophages, lymphocytes, and dendritic cells) and (v) an increased resistance to apoptosis [2]. The lung cancer cells preferentially metastasize to the brain, bones, liver and adrenal glands. The site of metastasis is influenced by histological subtype of cancer; small-cell carcinoma of lung frequently metastasizes to liver and brain [3,4], adenocarcinoma metastasize to bone and brain [5], squamous cell carcinomas more frequently show local invasion into the thoracic wall [6,7]. The primary determinants of patient prognosis include tumor histology, tumor stage, acquired EMT phenotype and presence or absence of EGFR mutations [8]. This brings into focus the necessity to regulate the metastasis of different lung tumors and their underlying signaling mechanisms by repurposed drugs known to act on EMT pathway. TGF-β Transforming growth factor (TGF)-β is a multitasking cytokinewhich signals cell cycle arrest, apoptosis and EMT [9,10]. EMT of tumor cells, increases their motility and metastasis. Previously, the role of TGF-β in excessive tumor cell migration and invasion has been demonstrated in gliomas [11], esophageal squamous cell carcinoma [12]. Therefore we studied the Transforming Growth Factor (TGF)-β1/BMP/Smad signaling pathway in the development of EMT in lung adenocarcinoma cells. The Smads are named after the C.elegans Sma and Drosophila Mad [13] and are divided to three categories: (1) Receptor-regulated R-Smads-1,2,3,5,8/9: Smad-2/3 mainly mediate signaling from TGF-β subfamily while Smads-1/5/8 transduct signaling from bone morphogenic protein (BMP); (2) common-mediator Smad (co-Smad) which includes Smad-4, and serves as the central mediator of both TGF-β and BMP signaling pathway; and (3) inhibitory Smad (I-Smad): Smad-6,7 which inhibit R-Smad phosphorylation, prevent their complex formation with co-Smad and antagonize the TGF-β/BMP signaling activity in a feedback-regulatory manner [14]. Pirfenidone is an anti-fibrotic agent used for treatment of idiopathic pulmonary fibrosis. It inhibits TGF-β induced EMT by acting on multiple targets; cytokines such as TNF-alpha, [15] and growth factors-TGF-β [16]. However, the precise molecular activities of pirfenidone remain incompletely understood [17]. In malignancies, pirfenidone has been shown to inhibit the TGF-β expression in renal cell carcinomas [18], malignant gliomas [19], pancreatic cancers [20] and possibly serve as a possible adjunct to therapies by disrupting the associated desmoplasia. Thus, pirfenidone may have an anti-cancer effect in addition to an anti-fibrotic effect via inhibiting type 3 EMT. The common pathophysiological backgrounds of IPF and lung cancer, have suggested the possible mutual or combined use of anti-fibrotic and anti-neoplastic drugs to treat these highly lethal diseases [21]. In lung cancer, pirfenidone has been suggested to suppress tumor growth via its anti-proliferative activity and direct inhibitory effect on EMT in NSCLC cell [22]. Yamamoto et al in 2020 observed that pirfenidone might be safe first-line chemotherapeutic agent when combined with carboplatin or immune checkpoint inhibitor therapy in 14 lung cancer patients with IPF [23]. In the present study, we elaborate the differential targeting of the TGF-β-Smad-1-7 and EMT pathway in lung cancer by repurposed drug, pirfenidone.
2. Materials and Methods
2.1 Materials
A-549 cancer cell line (NCCS, Pune), Dulbecco's modified Eagle's medium (DMEM), cat no: Al 007a, Hi-Media, India),10% fetal bovine serum (Invitrogen), 6 well culture plates (Cat no. 353046, BD Biosciences, U.S.A), trypsin-EDTA solution (cat no: TCL048, Hi-Media Laboratories, Nashik, India), Bleocip (Bleomycin sulphate, Cipla Ltd), Pirfenidone (5-Methyl-1-phenyl-2-(1H)-pyridone, P2116, Sigma- Aldrich India Ltd) and Pirfenex, Cipla Ltd), MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide)(Cat No- TOX1-1KT), Anti Mouse and rabbit (ExtrAvidin® Peroxidase, Extra3 and Extra 2 respectively, Sigma Life Sciences, USA), NovaRED substrate kit (SK-4800 Vector labs, USA), Meyer’s Hematoxylin, AmbionTRIzol® (Invitrogen 15596018), Chloroform, isopropanol, MMLV (M0253S, New England Biolabs), Primers (Sigma-Aldrich, India, Table 1), RNase (M0314, New England Biolabs (NEB), dNTPs (N0447S, NEB), Random primers (S1330S, NEB), SyBr Green (S4438 Jumpstart®Taq Ready Mix, Sigma Life Sciences). All reagents were of the highest commercial grade available.
2.2 Cell Culture
The human adenocarcinoma cell line (A-549) was purchased from the NCCS, Pune and maintained a humidified 5% CO2 atmosphere in CO2 incubator (New Brunswick Inc Scientific Co., U.S.A.) at 37°C in Dulbecco’s modified Eagle Medium (DMEM), containing 10% FBS which was supplemented with 100 U/ ml Penicillin, Streptomycin (100 μg/ml), and non-essential amino acids, in T-25 flasks (BD Biosciences, U.S.A). Subcultures of A549 cells were seeded in 60-mm culture dishes to analyze TGF-β, Smad-1-7, Vimentin, E-Cadherin mRNA expression. Cells were seeded at a density of 2–3×104 cells / ml and passaged when grown to 70-80% confluence. Subcultures of A-549 cells were done into 6 well culture plates at a density of 1× 105cells/well. Cells were passaged by detaching the cells with trypsin-EDTA solution [24,25]. When the cells reached 70-80% confluence, the medium was replaced with serum-free DMEM. Cells were subsequently stimulated for 4, 6, 8, 24, and 48h with: (a) culture medium alone Group-I- A549 cells (Saline control), (b) Group-II- A549 cells + bleomycin (50mM), (c) GroupIII-A549 cells+Bleomycin+Pirfenidone (500µg/ml). Cells were harvested at different time intervals- 4,6,8,24,48 hours and TGFβ1, SMAD-1-7, Vimentin, E-Cadherin mRNA levels were evaluated at all intervals.
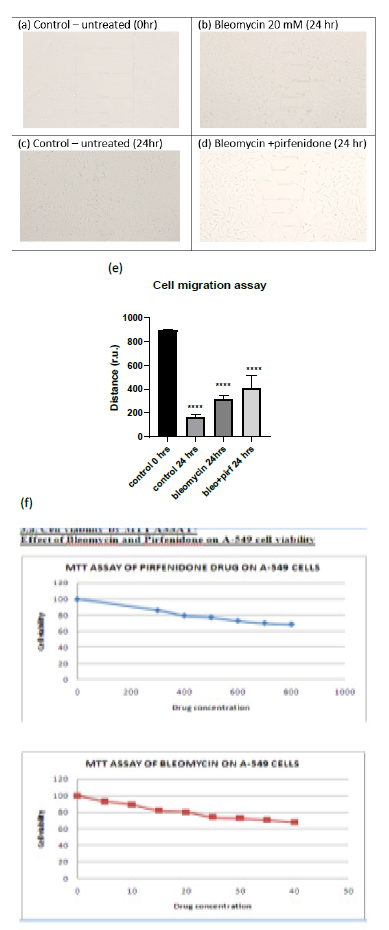
2.2.1 Cell migration Assay: The cell motility and adhesion in vitro were assessed by two-dimensional wound healing assay using the traditional pipette tip-scratch assay technique (scratch-wound assay). A-549 cells were cultured in DMEM in sterile 100 mm culture dishes/ 6-well plates (Nunc) and then serum-deprived for 24 h for 3 days to a confluent monolayer (at 37?C and 5% CO2). After cells reach confluence, a wound is made by scratching the cell monolayer in a straight line with a sterile P-200 pipette tip, in a straight line in the center of the dish to only detach central cells. The plates were then washed twice with 1ml of sterile 1X PBS to remove the floating cells and debris and incubated with 2ml of incomplete DMEM supplemented with bleomycin (50mM), pirfenidone (500µg/ml), and combination of the two agents (Bleomycin+Pirfenidone). Images were acquired at initial (0 h) and at 24 hours after drug therapy and the length of the wound area (r.u.) between one side of the scratch and the other was measured. The change in wound area (the distance cover by the cells in the wound closure) was then measured in at least 4 areas of each group (n=12) and was analyzed with Image J software (https://imagej.nih.gov/ij/ National Institute of Health NIH, Bethesda, MD, USA). Cells treated with drug-free medium were considered as controls [26].
2.2.2 Cell Viability and Proliferation: This was assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay using 96-well plates at a seeding density 105 cells per mL After seeding, the cells were allowed to attach in a CO2 incubator at 37 ºC for 24 h. After 24 hours, the culture media was removed and replaced with100 mL of fresh complete medium containing bleomycin, pirfenidone and incubated for 24 h to observe the cytotoxic and proliferative effects. Briefly, 20 μL of MTT solution was added to each well (5 mg MTT mL-1) was dissolved in 1X PBS and syringe filtered it and incubated for 3 h at 37ºC for intracellular formazan crystals formation. Then, 100 μL (0.1% v/v) of DMSO was added to each well to dissolve the formazan crystals. After 5 min of shaking, the optical density (OD) at 570 nm was measured using monochromator (biiotek instrument Epoch 2 microplate reader). The cell viability after bleomycin (20 mM) and after pirfenidone (500 µg/ml) was assessed in A549 cell line at 24 h.
2.3 RNA Extraction and Quantitative Real Time PCR
Total RNA was extracted from cultured cell homogenates using the guanidium thiocyanate- phenol-chloroform method. Samples were digested with DNase I (Qiagen) to remove contaminating genomic DNA. RNA was precipitated with 500 μl isopropanol (v/v), washed with 75% ethanol and reverse transcribed to cDNA using the MMLV-RT enzyme and random hexamers. The cDNA was amplified with the cycling condition: PCR activation at 95?C for 5 mins, 35 cycles of denaturation at 95?C for 30 secs, annealing at 60??C for 35 secs, and extension at 72?C for 30 secs and a final extension at 72?C for 7 mins. qRTPCR reactions were performed in 72-well Rotar-Gene Q (Qiagen) using SYBR Green PCR Master Mix and specific sequence primers (Sigma). Beta-actin mRNA amplified from the same samples was the internal control. The primers used were synthesized by Sigma-Aldrich, India as per Table 1. The relative expression of each targeted gene was normalized by subtracting the corresponding housekeeping genes (β-actin) threshold cycle (Ct) value using the comparative CT method (??Ct method).
2.4 Statistics
All values are expressed as means ± SEM. The minimum number of replicates for all measurements was at least 3. Differences between multiple groups were compared by one-way analysis of variance. Neuman keule’s test was used as post-hoc test. Significance was assumed at p < 0.05.
3. Results
3.1 Cell Viability and Migration Assay
Cell migration is a key procedure involved in physiological and pathological processes such as embryological development, tissue formation, immunological response, inflammation, and cancer progression [27]. Individual cancer cell migration is mediated by cytoskeletal activity in the cells in the early stages of invasion/ metastatic process and is similar to in vivo physiological developmental processes [28]. In the present study we assessed the cell viability after pirfenidone and bleomycin by MTT assay and observed that 80% of A-549 cells were viable after treatment with 600 µg/ml pirfenidone and 20 mM bleomycin respectively (Figure 3.1-f). Further, we assessed the two-dimensional wound healing assay using the traditional pipette tip-scratch assay technique to assess the cell motility and adhesion in vitro. The mean distance of the wound length (r.u.) was estimated as a measure of wound closure by the proliferating cells and mean±SEM recorded. As compared to control group, a significant reduction in wound area was seen after bleomycin induced EMT (Figure 3.1-e). Pirfenidone treatment to the bleomycin treated cells revealed an increase in wound area indicative of reduced EMT and migration potential of the A-549 cancer cells after pirfenidone therapy (Figure 3.1-e).
Figure 3.1: (a-d) Analysis of A-549 cell migration by in vitro wound healing assay. (e) Distance of wound closure. Note the significant reduction in wound length indicative of EMT induced by bleomycin and its increase after pirfenidone therapy. (f) Cell viability assay by MTT after pirfenidone and bleomycin treatment.
3.2 TGF-β1gene Expression
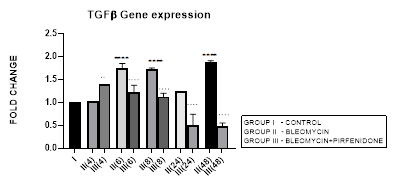
3.2.1 TGF-β1 Expression in Bleomycin treated A549 cells: TGF-β1 has pleiotropic action on cell behavior [29]. It induces fibroblast proliferation [30,31] and transformation via the canonical and non-canonical TGF-β signaling pathway [32]. In tumors, TGF-β signaling plays a dual role by helping the tumor cell to evade immune system recognition and increasing their risk of recurrence and metastasis [33]. While simultaneously inhibiting the proliferation of normal epithelial cells and hematopoietic cells [31,34]. TGF-β along with its related genes such as BMPs, activin/inhibin, influences stromal phenotypes and results in cancer progression [35,36]. In NSCLC, TGF-β upregulation has shown to significantly predict poor prognosis and survival [37]. In the present study, we demonstrate upregulated TGF-ß1 expression after bleomycin treatment of A549 cells (Figure 3.2). This is associated with change in AEC morphology to fibroblast-like cells as compared to the control group in which the classic cobblestone epithelial morphology is maintained. This is similar to previous study by Chen et al, 2016 who demonstrated change in AEC morphology to fibroblast-like cells after treatment with 20 μg/mL bleomycin.
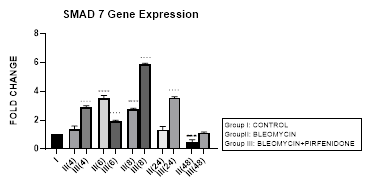
Figure 3.2: Bimodal upregulation in TGF-ß1 gene expression at 6,8 and 48 hrs after bleomycin treatment. Note the significant reduction of TGF-ß1 expression and attenuation of EMT after pirfenidone therapy. P<0.0001**** I vs. II (6),II(8),II(48); P<0.0001….II(6)vs.III(6), II(8)vs.III(8), II(24)vs.III(24), II(48)vs.III(48); P<0.01..II(4) vs III(4).
3.2.2 Pirfenidone Down regulates TGF-β1 and Reverses the EMT of AECs: Pirfenidone acts as an antifibrotic by; (i) suppressing macrophage-driven cytokines such as IL-1, TNF-α, TGF-β1, PDGF, HSP47, MCP-1 [17] (Ruwanpura, 2020), (ii) attenuating IL-6 signaling [38] (ii) reducing the numbers of infiltrating macrophages and suppressing M2 macrophage polarization [39], (iii) reducing TGF-β1 expression and inhibiting TGF-β1-induced collagen synthesis [24,40-42], (iv) suppressing fibroblast proliferation and fibroblast differentiation into myofibroblasts by attenuating TGF-β-mediated Smad-3 phosphorylation, Wnt/GSK-3β/β-catenin and Smad-2/3 signaling [43,44]. In NSCLC, pirfenidone inhibits the extracellular matrix deposition and tumor-stroma interaction [45,46]. In present study, we demonstrate the efficacy of pirfenidone in inhibiting TGF−β1 expression and its differential action on Smad-1-7 downstream signaling to reverse the EMT and migration potential of the A-549 carcinoma cells (Figure 3.1-a-f, 3.2). Thus indicative of its potential in reducing lung cancer metastasis [22].
3.3 Smad-1 Gene Expression
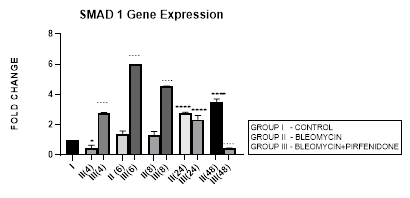
3.3.1 Smad-1 Gene Expression in Bleomycin treated A549 cells: The Smad-1 is an inhibitory Smad that activates BMP pathway along with Smads-5 and 8 [35,47] and inhibits TGF-β1 mediated fibrotic gene expression and fibrosis [48]. In present study, suppression of Smad-1 expression was seen in early phase (up to 8 hours) which correlated with TGF-β upregulation in bleomycin treated A549 cells. The Smad-1 gene expression progressively increased at 24 and 48 hrs (Figure 3.3) however inspite of the increase of Smad-1, TGF-β1 upregulation and progression of EMT was observed at 48 hours after bleomycin therapy (Figure 3.2). This is explained by the TGF-β1-induced Smad-1 upregulation and phosphorylation that occurs via two classes of type I receptors; TGFBR1 and ACVR1. TGFBR1 phosphorylates and activates ACVR1, which in turn phosphorylates Smad-1 [29].
Figure 3.3: Increase in Smad-1 gene expression in bleomycin treated A549 cells at 24 and 48 hrs. Note the significant upregulation of Smad-1 after pirfenidone therapy at 6,8 and 24 hrs and reduction in late phase (at 48 hours). P<0.0001**** I vs.II(24), II(48), III(4), III(6),III(8); P<0.0001….II(4) vs. III(4), II(6) vs. III(6) ,II(8) vs. III(8), II(48) vs. III(48).
3.3.2 Pirfenidone upregulates Smad-1: So far studies have mainly focused on pirfenidone’s inhibitory effect on TGF-β1 induced rSmad proteins-2,3 [49] and their downstream signaling via MUC1-CT phosphorylation, β-catenin activation, nuclear complex formation of phospho-SMAD3/MUC1-CT/active β-catenin [50]. Recent studies have suggested that it is the impairment of the antifibrotic TGF-β-Smad1/5/8/9 pathway that correlates with enhanced EMT [51]. The activation of antifibrotic iSmad signaling via BMP7-mimetic compounds (small-molecules that activate the TGF-β-Smad1/5/8/9 pathway) [51] has shown attenuation of EMT in mouse model of chronic renal injury [52].In present study, we demonstrate the efficacy of pirfenidone in significant upregulation of Smad-1 expression by A-549 cells at 6 and 8 hrs (Figure 3.3) that correlates with reduction of TGF-β1 and attenuation of EMT. Our findings show for the first time that pirfenidone induced activationof iSmad-1 from early phase is one of its important signaling mechanism in reversing EMT in lung adenocarcinoma cell line.
3.4 Smad-2 Gene Expression
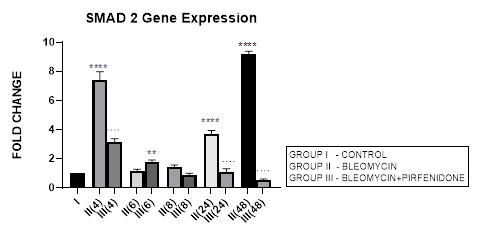
3.4.1 Smad-2 Gene Expression in Bleomycin treated A549 cells: Lung injury caused by bleomycin induces NF-κB expression and activates TGF-β1 binding to its receptors TβR-I, II resulting in Smad-2 phosphorylation. The phosphorylated Smad-2 forms heterooligomeric complex with Smad-3 and 4 and translocates to the nucleus. This initiates the canonical TGF-β1-Smad-2 signaling pathway [53] and induces target gene transcription [54]. The Smad-2 signaling pathway plays an important role in the pathogenesis of remodeling and fibrosis [55] and predominantly regulates genes during development [56]. Thus the TGF-β1/Smad-2 pathway links the inflammatory (NF-kB) and oxidative stress (Nrf2-NOX4) to progressive lung fibrosis and can serve as a key point for its treatment [53]. In lung cancer, Zeng et al, 2020 report the distinct expression and prognostic value of individualSmadsin patients with NSCLC. They report significant down-regulation of Smad-6/7/9mRNA levels and aberrantSmad-2/3/4/5/6/7/9mRNA levels in NSCLC to correlate with prognosis of NSCLC and serve as potential targets for NSCLC and prognostic biomarkers. In present study, bleomycin treated A-549 cells showed TGF-β upregulation resulting in bimodal increase in Smad-2 gene expression in at 4, 24 and 48 hrs and EMT of A-549 cells (Figure 3.4, 3.13). The rSmad-2 and 3 localise to the nuclei of the infiltrating macrophages and to type II epithelial cells and are associated with progression of fibrosis from day 7 to 14 [57].
3.4.2 Pirfenidone downregulates Smad-2: Previously Choi et al, 2012, have observed that pirfenidone blocks TGF-β signaling by preventing nuclear accumulation of active Smad2/3 complexes rather than phosphorylation of Smad2/3. This can have therapeutic implications. Recently Liu et al, 2019, have linked the NF-kB pathway with the TGF-β1-Smad2/ NOX4-Nrf2 signaling pathways and suggested the role of anti-inflammatory and anti-oxidation drugs as a new direction of treatment of lung fibrosis [55]. In the present study we demonstrate the significant down regulation of Smad-2 expression from early phase after pirfenidone therapy that resulted in reversal of EMT of A-549 cells (Figure 3.4, 3.13).
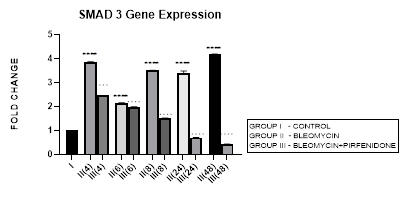
3.5 Smad-3 Gene Expression
3.5.1 Smad-3 Gene Expression in Bleomycin treated A549 cells: In present study we elaborate the time course of TGF-β signaling and Smad-2, 3 regulations in bleomycin treated A549 cells. Bleomycin treated A-549 cells showed a persistent upregulation in Smad-3 gene expression in present study (Figure 3.5). Smad-3 expression differed from Smad-2 which showed a bimodal upregulation after bleomycin (Figure 3.4). Thus bringing into focus the fact that, while both Smads 2 and 3 mediate signals from TGF-β and activin, these two Smads clearly have differential time course of their expression (Figure 3.4, 3.5, 3.12) and nonredundant functions [58]. Smad-3 binds to DNA through its MH1 domain and directly activates TGF-β1, c-fos, and Smad-7 genes while negatively regulating the MMP-1 promoter [59]. This is unlike Smad-2, which activates transcription indirectly by binding to transcription factors. TGF-β-mediated induction of MMP-2 and activation of the activin-response element Lux reporter are Smad-2 dependent [59]. Thus, suggesting specific roles and mechanisms for Smads-2 and 3 in signaling [59]. The collagen gene promoters induced by TGF-β1-Smad-3 include COL1A1, COL1A2, COL3A1, COL5A2, COL6A1 and COL6A3 [58,60]. Their serum levels may be useful biomarkers for idiopathic pulmonary fibrosis [61]. Smad-3 mediates progression of fibrosis by; (1) promoting epithelial to mesenchymal transformation, (2) recruitment of inflammatory cells and fibroblasts, (3) auto induction of TGF-β in the AECs (4) induction of collagen synthesis. Smad-3 is therefore considered as a potential target in the treatment of fibrosis-related diseases [62]. Chemically synthesized miRNA inhibitors of Smad-3 are being explored (miRNA 326) [63,64]. The Smad-3 inhibition interferes with expression of profibrotic genes (collagens, PAI-1, TIMP-1) and inhibits fibrosis [58].
Figure 3.5: Persistent upregulation in Smad-3 Gene expression in bleomycin treated A549 AEC’s. Note the significant reduction in Smad-3 by pirfenidone treatment. P<0.0001 **** I vs. II(4),II(6),II(8),II(24),II(48); P<0.0001….II(4)vs.III(4), II(6)vs.III(6), II(8)vs.III(8), II(24)vs.III(24), II(48)vs.III(48).
3.5.2 Pirfenidone downregulates Smad-3: We demonstrate a significant reduction in Smad-3 expression of A-549 cells after pirfenidone treatment. Smad-3 levels reduced at all-time intervals from early phase (4 hrs), to below baseline levels at 24 and 48 hours (Figure 3.5). This correlated with attenuation of EMT (Figure 3.10 and 3.11). This is similar to pirfenidone attenuation of Smad-3 phosphorylation and TGFβ1/Smad3-induced signaling resulting in suppression of TGF-β-mediated fibroblast proliferation and fibroblast-myofibroblast differentiation in radiation-induced lung fibrosis [43]. This explains why Smad-3 null mice are resistant to bleomycin-induced pulmonary fibrosis, radiation-induced cutaneous fibrosis, carbon tetrachloride-induced hepatic fibrosis and streptozotocin induced glomerular fibrosis [58]. Smad-3 inhibitors such as 1,25-dihydroxyvitamin D3, have shown suppression of TGF-β levels [58,65]. However, long-term partial suppression of TGF-β signaling through Smad-3 inhibition may lead to potentially serious adverse effects; (i) may stimulate the development of autoimmune diseases. (ii) May increase expression of iSmad-7 protein [66]. To avoid these and similar potential complications, fibrotic conditions in which Smad-3 inhibitors can be applied locally have been advocated. Surprisingly, phosphorylated Smad-3 was unchanged in lung tissue of patients with IPF treated with pirfenidone, a discrepancy possibly due to small patient numbers and timing of tissue collection [17].
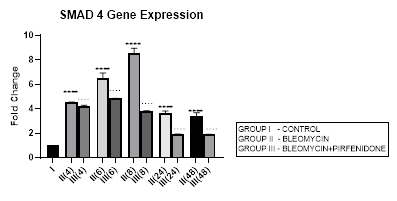
3.6 Smad-4 Gene Expression
3.6.1 Smad-4 Gene Expression in Bleomycin treated A549 cells: Smad-4, is the central mediator of TGF-β and BMP signaling. It forms a heterooligomeric complex with Smad-2 and Smad-3 and translocates them to the nucleus to induce target gene transcription and downstream TGF-β1 activities. In present study, bleomycin treated A-549 cells showed a persistent up regulation in Smad-4 gene expression with maximum up-regulation at 6 and 8 hour (Early phase), (Figure 3.6, 3.13). This was associated with upregulation of TGF- β1 (early phase- 6 and 8 hour) (Figure 3.2, 3.13) and indicative of its role in regulating canonical TGF-β signaling. This action of Smad-4 is through a feedback mechanism involving inhibition of miRNA-425 and downregulating TGFBR2 receptor [67]. Other miRNAs that regulate TGFBR2 expression, include miR-21, 145, 211, 130a-3p, and miR-17∼92 family clusters [67,68]. The Smad-4-dependent TGF-target genes, at the mRNA level, are, PAI-1, Smad-7, AKAP12, and ITGB6. The TGF induced- Smad-4 independent genes include TMEPAI, CDKN1A (p21), CDKN2B (p15), SNAI2 (Slug), and c-JUN. Bleomycin also induced upregulation and nuclear localization of Smad-2/3/4 complex in A-549 cells and promoted expression of pro-apoptotic genes, such as TIEG (TGF-β-inducible early response gene), DAPK (the death-associated protein kinase), GADD45β and Bim [69]. Thus Smad-4 has been suggested as a prognostic indicator in cancer and needs to be explored [69].
Figure 3.6: Persistent up regulation in Smad-4 gene expression in bleomycin treated A549 cells from 4 to 48 hrs with maximum up regulation seen at 8 hour interval. Pirfenidone therapy significantly downregulated Smad-4 at all-time intervals with maximum reduction at 24 and 48 hours. However the levels did not reach up to baseline levels even at 48 hours. P<0.0001**** I vs. II(4),II(6),III(6),III(8),II(48),III(24); P<0.0001….II(6)vsIII(6), II(24) vs III(24) , II(48) vs III(48).
3.6.2 Pirfenidone downregulates Smad-4: The canonical TGF-β/Smad-4 signaling pathway plays, (i) a tumor suppressive role in early-stage cancer (ii) induces cell cycle arrest and apoptosis by acting at the G1/S checkpoint to make cells stay at the G1 phase [70], (iii) Smad-2/3/4 form a complex with transcriptional factor FoxO to promote transcription of p15 and p21. P21 acts both as a tumor-suppressor gene and an inhibitor of apoptosis [71]. In present study, pirfenidone therapy significantly downregulated Smad-4 at all-time intervals with maximum reduction at 24 and 48 hours. However the levels did not reach up to baseline levels even at 48 hours (Figure 3.6). This was associated with reduction in TGF-ß1 gene expression (Figure 3.2) and reversal of EMT (Figure 3.10, 3.11) in a Smad-4 dependent manner. However, Smad-4 loss can reduce the apoptosis of the alveolar epithelial cells and promote tumor progression initiated by other genes and prove to be a double edged sword [69].
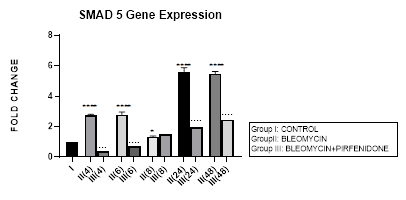
3.7 Smad-5 Gene Expression
3.7.1 Smad-5 Gene Expression in Bleomycin treated A549 cells: In present study, bimodal increase in Smad-5 Gene expression is seen in bleomycin treated A549 cells at early phase (4,6 hrs) and late phase (24,48 hours) (Figure 3.7, 3.13). An associated upregulation of TGF- β1 was seen at 6 and 8 hour interval (Figure 3.2). Indicative of role of TGF-β in activating Smad-5 and inducing its phosphorylation [29]. This increase in Smads activates the bone morphogenetic protein (BMP) pathway [35,47] and BMP-2/4 in turn transmits signals via Smad 1, 5, and 8 [72]. BMP-7 binds to the BMPIIRs to (i) activate R-Smad1/5/8 in a positive feedback loop [73] (ii) activate the activin-like kinase receptors (ALK2, ALK6, ALK3) and (iii) inhibit TGF-β1 mediated fibrotic gene expression and fibrosis [48]. The TGF-β/Smads and the BMP/Smads thus counter-regulate each other to maintain the balance between two pathways in the pathophysiological process. However, a subsequent study has failed to show this counter-regulating activity [74], suggesting the high level of complexity between the TGF-β/Smad and BMP/Smad pathways under disease conditions and the need for their elaboration [75].
Figure 3.7: Bimodal increase in Inhibitory Smad-5 Gene expression in bleomycin treated A549 cells at 4 and 6 hours and then at 24 and 48 hours. Note the significant down-regulation of Smad-5 after pirfenidone therapy at 24 and 48 hours, which however remains above baseline levels and is responsible for reversal of fibrosis. P<0.0001**** I vs. II(4),II(6),II(24),II(48); P<0.1* I vsII(8) P<0.0001; ….II(4)vsIII(4), II(6)vsIII(6), II(24) vs III(24), II(48) vs III(48).
3.7.2 Pirfenidone downregulates Smad-5: In the present study, significant downregulation of Smad-5 occurred after pirfenidone therapy at 24 and 48 hours. However, even at 24 and 48 hours, the Smad-5 levels remained above baseline levels (Figure 3.7). Thus indicative of the Smad-5/BMP signaling and its protection against bleomycin induced EMT that is responsible for reversal of fibrosis. This is similar to protective action of enhanced endogenous BMP pathway in attenuating bleomycin induced lung fibrosis in mice model [76]. The influence of the Smad dependent BMP signaling is being studied in bone remodeling [77]. We demonstrate, in this study the effect of Pirfenidone in modulating Smad-5 and TGF-β and BMP pathway in lung adenocarcinoma cells.
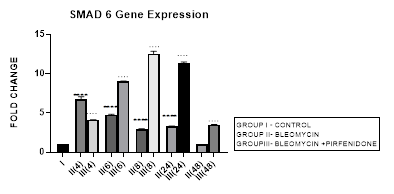
3.8 Smad-6 Gene Expression
3.8.1 Smad-6 Gene Expression in Bleomycin treated A549 cells: The I-Smads-6,7 are located in the nucleus of resting cells. When stimulated by TGF-β they enter the cytoplasm (Chen et al., 2002) and bind to type I receptors. iSmads competitively interfere with recruitment and phosphorylation of R-Smads, and inhibit the activity of R-Smads and formation of their Co-Smad complexes [78]. In the present study, we demonstrate the peak upregulation of Smad-6 at 4 hrs followed by its progressive downregulation up to 48 hours after bleomycin (Figure 3.8). This reduction of inhibitory Smad-6 was associated with progressive upregulation of rSmads-2,5 and EMT of bleomycin treated A-549 cells (Figure 3.10, 3.11, 3.12, 3.13). The upregulated iSmad-6, (i) activates the BMP pathway and prevents recruitment and phosphorylation of receptor (R)-Smads. (ii) The I-Smad-6 brings the E3 ubiquitin ligases Smurfs 1 and 2 (Smad ubiquitination regulatory factors 1 and 2) to the Type I receptor which subsequently ubiquitinate and degrade the R-Smads [58]. The loss of R-Smads 2,3 interferes with TGF-β-mediated induction of EMT and down regulates genes for collagens, PAI-1 and TIMP. Suggesting that, small molecule up-regulators of Smad-6 and inhibitors of Smad 2,3 may have potential in the treatment of pathological fibrotic diseases.
Figure 3.8: Peak increase in inhibitory Smad-6 gene expression at 4hours after bleomycin treatment followed by its downregulation up to 48 hours. Note the significant up-regulation of Smad-6 after pirfenidone therapy at all time intervals up to 48 hours.P<0.0001**** I vs. II(4), II(6), II(8), II(24); P<0.0001….II(4)vs. III(4), II(6)vsIII(6),II(8)vsIII(8), II(24) vs III(24) , II(48) vs III(48).
3.8.2 Pirfenidone up regulates Smad-6: In the present study, we demonstrate significant upregulation of the Smad-6 expression by pirfenidone at all-time intervals up to 48 hours. This was accompanied by suppression of the TGFβ1/Smad induced EMT of A-549 cells and their differentiation into myofibroblasts (Figure 3.10, 3.11). The Smad-6 inhibits activation of R-Smads by Type I receptor kinases via the BMP pathway while the Smad-7 inhibits activation of R-Smads via the TGF-β/activin pathway [58].Thus suggesting, novel role for Smad-6,7 in aberrant parenchymal remodelling during lung development, injury, and repair. Previous studies in embryogenesis models have demonstrated the differential regulation of lung branching morphogenesis and cytodifferentiation by Smad-7, but not by Smad-6 [79]. However in adult lung, an increased expression of Smad-6 has been shown to be involved in the regulation of fibrotic conditions [80]. Thus targeting SMAD6 can be used as a strategy to inhibit the phosphorylation of SMAD2/3, and thereby block TGF-β signaling [81]. Using this strategy, Sharma et al, 2011 have advocated the utility of pirfenidone in treating renal fibrosis and diabetic nephropathy in animal model [82].
3.9 Smad-7 Gene Expression
3.9.1 Smad-7 Gene Expression in Bleomycin treated A549 cells: In present study, an early upregulation of inhibitory Smad-7 is seen at 6, 8 hrs after bleomycin followed by its progressive downregulation up to 48 hours (Figure 3.9, 3.12). An upregulated Smad-7 initially inhibits activation of R-Smads by blocking promoter transactivation by TGF-β in fibroblasts [58]. The Smad-7 also binds to the E3 ubiquitin ligases Smurfs 1 and 2 and degrades the R-Smads [58]. The subsequent reduction of inhibitory Smad-7, is associated with progressive EMT and fibrosis. This is similar to previous study, which demonstrated suppression of type I precollagen mRNA, hydroxyproline, and attenuation of bleomycin-induced lung fibrosis in C57BL/6 mice by exogenous Smad-7, but not by Smad-6 [83]. Also in the present study, the upregulation of I-Smad-7 at 6 and 8 hour correlated with maximal upregulation of Smad-4 at the same time (Figure 3.6). This correlation has indicated the Smad4-dependent contribution in activation of Smad-7 [84]. However, this is a small contribution and majorly Smad-4 signaling is not considered essential for TGFβ-induced Smad-7 expression [85].
Figure 3.9: An upregulation of inhibitory Smad-7 after bleomycin is seen at 6, 8 hrs, followed by its progressive downregulation up to 48 hours. This reduction of inhibitory Smad-7, is associated with progressive EMT. Note the significant upregulation of Smad-7 after pirfenidone therapy at all time intervals up to 48 hoursresulting in inhibition of TGF pathway. P<0.0001 **** I vsII(6), II(8), P<0.001 *** I vs II(48), P<0.0001 .... II(6) vs III(6), II(8) vs III(8), II(24) vs III(24), P<0.0001 ....II(48) vs III(48).
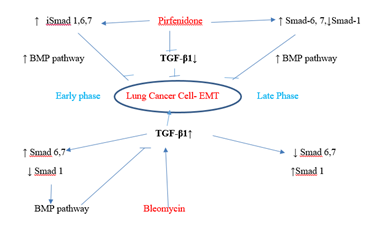
Figure 3.12: Pirfenidone action on TGF- β1- iSmad-1,6,7 signaling pathways. In, A-549 cells derived from lung adenocarcinoma, bleomycin impaired the antifibrotic TGF-β-Smad-1 (Early Phase) and iSmad-6, 7 (Late phase) pathway and induced EMT. Pirfenidone acts by downregulating TGF-β1 and upregulating iSmads- 1 (Early phase), iSmad-6,7 (All phases).
3.9.2 Pirfenidone up regulates Smad-7: Recently, the upregulation of Smad-7 is being investigated with renewed interest; (i) Newly identified molecule, RS4651 has been shown to upregulate Smad-7 expression and inhibit the proliferation and migration of MRC-5cells (Shirong, 2021). (ii) similarly, knocking down miR-182-5p has been shown to effectively upregulate Smad-7 expression and inhibit lung fibrosis. Suggestive of miR-182-5p inhibition to be a promising therapeutic approach in treating IPF [86] (Chen, 2020). (iii) miR-326 causes significant downregulation of TGF-β1, Smad-3, matrix metalloproteinase-9 (MMP-9), and upregulates Smad7, which in turn have good anti-fibrosis effects. In present study, we demonstrate the efficacy of pirfenidone in significantly upregulating Smad-7 at all-time intervals up to 48 hours (Figure 3.9, 3.13) resulting in inhibition of TGF pathway (Figure 3.2) and attenuation of EMT of A-549 lung adenocarcinoma cells. Previously, the efficacy of pirfenidone has been demonstrated in decreasing miR-21 expression and increasing Smad7 mRNA expression levels and then inhibiting the TGF-β1/Smad3 signaling pathway [87] (Bi, 2021).
↓ rSmad-2,3 Pirfenidone↓ Smad-2,3,5
↓ Smad-4↓ Smad-4
TGF-β1↓
Early phaseLung Cancer Cell- EMTLate Phase
TGF-β1↑
↑Smad-2,3,5↑Smad-2,3,5
↑Smad-4↑Smad-4
BMP pathwayBleomycin
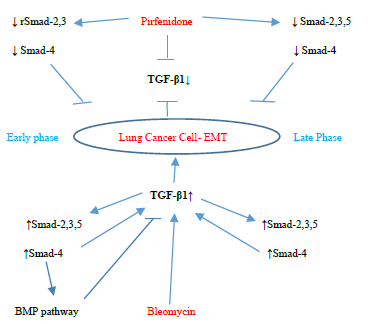
Figure 3.13: Pirfenidone action on TGF- β1, rSmad- 2,3,5 and coSmad 4 signaling pathways. In, A-549 cells derived from lung adenocarcinoma, bleomycin upregulated the profibrotic TGF-β-rSmads-2,3,5 and coSmad-4 pathway from the Early Phase and induced EMT. Bleomycin differentially regulated Smad-2 and 3, with bimodal upregulation of Smad-2, while Smad-3 was persistently upregulated. Pirfenidone acts by downregulating TGF-β1 and Smads- 2,3,4 (from Early phase) and Smads-2,3,4,5 (Late phase).
3.10 E-cadherin Gene Expression
3.10.1 E-cadherin Gene Expression in Bleomycin treated A549 cells: EMT is characterized by gradual conversion of alveolar epithelial cells to elongated fusiform mesenchyme cells resulting in the down-regulation of epithelial markers (E-cadherin) and the up-regulation of mesenchymal markers (N-cadherin, Vimentin, Fibronectin, α-SMA) [88]. E-cadherin normally localizes at the plasma membrane [89] and its cytoplasmic relocation, downregulation or loss in pluripotent stem cells can trigger EMT [90]. In addition, E-cadherin is also an important tumor-suppressor gene. In cancer cells, loss of E-cadherin gene expression causes dysfunction of the cell–cell junction system, triggering cancer invasion and metastasis. Therefore, we assessed the E-cadherin expression after bleomycin treatment in A-549 cells. In present study, a progressive decrease in E-cadherin gene expression from early phase up to 48 hours, indicative of loss of epithelial characteristic was seen after bleomycin instillation (Figure 3.10). This correlated with reduction of number of cancer calls as also with increasing EMT and migratory capacity of the surviving cells (Figure 1.b,e). The acquired mesenchymal phenotype has been shown to be mediated by activation of transcription factors; SNAI1, SNAI2 and ZEB1/2 [91] and manifests as enhanced migratory activity, extracellular matrix (ECM) production, invasiveness, and elevated resistance to apoptosis [92]. Thus enabling the cells to penetrate into blood vessels and lymphatics and disseminate to distant organs, where they form metastatic lesions [93].
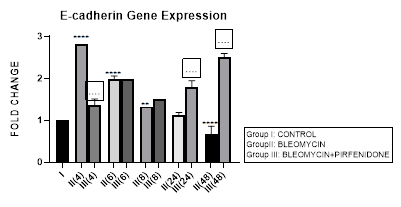
Figure 3.10: Progressive decrease in E-Cadherin gene expression in bleomycin treated A549 cells from early phase up to 48 hours indicative of loss of epithelial characteristic. Note the significant upregulation of E-cadherin after pirfenidone therapy from early phase up to 48 hours indicative of restoration of epithelial phenotype. P<0.0001****I vs. II(4),II(6),III(6),III(8),III(24),III(48); P<0.001***I vs. III(4); P<0.01**I vs. II(8),II(48); P<0.0001....II(4) vs III(4), II(24) vs. III(48), II(48) vs. III(48).
3.10.2 Pirfenidone up- regulates E-cadherin: In the present study, we demonstrate the significant upregulation of E-cadherin after pirfenidone therapy from early phase up to 48 hours indicative of its efficacy in restoration of epithelial phenotype. This is unlike previous study where treatment with pirfenidone alone did not significantly affect the E-cadherin expression while it successfully inhibited fibroblast migration [26]. Earlier also it was suggested that pirfenidone can only partially inhibit EMT changes [24] and E-cadherin down-regulation induced by TGF- was not attenuated by pirfenidone. Hisatomi et al. had reported that pirfenidone could inhibit collagen type I and heat shock protein 47 expression but not E-cadherin expression. They observed re-localization of E-cadherin to cytoplasm or nucleus in A549 cells after TGF-1 stimulation, and E-cadherin remained at the membrane following addition of Pirfenidone. This may in part explain why Pirfenidone was not found to alleviate TGF- induced downregulation of E-cadherin in their study. Pirfenidone is a pleiotropic molecule that inhibits TGF−β induced AEC-II to mesenchymal and fibroblast to myofibroblast transitions [50], fibroblast proliferation and mediates tissue repair in lung, liver and kidney tissue [24,40,42]. Several molecular mechanisms responsible for the anti-fibrotic action of pirfenidone are still being explored. Targeting, fibroblast activation, myofibroblast transition and tumor-stroma interaction in non-small cell lung cancer by pirfenidone may serve as a novel treatment strategy [38].
3.11 Vimentin Expression
3.11.1 Vimentin Gene Expression in Bleomycin treated A549 cells: In present study we demonstrate increase in vimentin gene expression from early phase up to 48 hours after bleomycin induced TGF-β in A549 cells indicative of gain of mesenchymal characteristic and EMT. Occurrence of EMT is also characterized by changes in cell morphology, isoform switching of the fibroblast growth factor receptor 2 (FGFR2) by alternative splicing [94]. In cancer, EMT is associated with invasion, metastasis and chemoresistance [95]. Thus, EMT relates to both organ fibrosis and malignant behavior of cancer. EMT is classified into three different subtypes: type 1-seen in implantation or embryo formation, type 2- occurs in wound healing and organ fibrosis, and type 3 seen in cancer[96] cells, which increases their invasion and metastasis to other tissues. Thus, finding an effective way to inhibit Vimentin and mesenchymal gene expression and EMT should offer an attractive therapeutic strategy for controlling cancer.
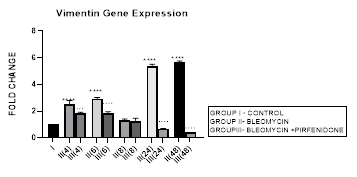
Figure 3.11: Bimodal increase in Vimentin Gene expression in bleomycin treated A549 cells from early phase up to 48 hours indicative of gain of mesenchymal characteristic. Note the significant downregulation of Vimentin after pirfenidone therapy at 24 and 48 hours to below baseline levels indicative of restoration of epithelial character. P<0.0001**** I vs. II(4), II(6),III(6), & II(24)&II(48); P<0.001… II(4) vs III(4); P<0.0001 ....II(6)vs III(6),II(24) vs III(24) , II(48) vs III(48).
3.11.2 Pirfenidone down-regulates Vimentin: Pirfenidone inhibits fibroblast activity, the crosstalk between cancer cells and fibroblasts [38], and can revert increased fibronectin, slug, PD-L1, and cell motility induced by TGF-β and FGF-2 [97]. In the present study we demonstrate the significant downregulation of vimentin after pirfenidone therapy at 24 and 48 hours to below baseline levels indicative of restoration of epithelial character. Thus, pirfenidone like nintedanib can induce EMT-reversion in human lung adenocarcinoma [95]. Pirfenidone may be a new therapeutic agent for adjuvant therapy in NSCLC patients to reduce cancer cell metastasis via regulation of EMT [22].
4. Discussion
Mortality rates in non-small cell lung cancer (NSCLC) patients remain excessively high despite the implementation of novel targeted therapies and chemotherapeutic regimes because of the tumor’s high metastatic ability [98]. The need to use repurposed drugs for inhibiting the epithelial-mesenchymal transition (EMT) in non-small cell lung cancer is critical. TGF-β is one such target as it is widely overexpressed in many cancers and its upregulation in tumors is related to poor prognosis, tumor vascularization and metastasis [99,100]. The TGF-β/Smad signaling pathway contributes to multiple steps of carcinogenesis, including; invasion, migration, mesenchymal transition and extracellular matrix synthesis [101]. Therefore, targeting the TGF-β/Smad pathway can function as an important tumor suppressor in cancers by inducing cell cycle arrest and reducing cell proliferation [101,102]. The downregulation of TGF-tumor suppressive response and loss of Smad-4 contributes to tumorigenesis [103] and may prove to be a double edged sword [69]. The TGFβ pathway has been targeted by using repurposed therapeutic approaches acting directly on the ligands, the receptors, canonical and non-canonical signaling and effectors as well as interacting pathways. Some of these approaches include; Neutralizing antibodies against TGFβs, (GC1008 (fresolimumab), inhibitory Proteins and Decoy Receptors (decorin), BMP-1/BMP-1/Tolloid-like proteinase (Tolloids) inhibition, inhibition of integrins to block TGFβ activation, ALK4/5 and type 2 TGFβRII receptor inhibition [104]. A common challenge for all these inhibitory approaches is the pleiotropic action of TGF-β. Many of these approaches have failed despite promising preclinical data due to unfavorable risk–benefit profiles in patients [104]. The Smads play a significant role in tumor progression and are another attractive target. Of the Smads, Smad-4 mutation is considered by some to be the most common protein instability in tumors and plays a crucial role in tumor suppression [99,100]. While Smad-4, has been shown to affect Smad-2 phosphorylation and increase Smad-2 auto-inhibition and suppress tumor growth [69,100]. Other groups do not considered Smad-4 to be involved in the TGF-induced EMT, tumor promotion [84] and metastasis [105]. Some studies have suggested the existence of two populations of TGF-β target genes distinguished by their dependency on Smad-4 and having diverse effect on tumor growth [84]. Similarly, Loss of Smad3 expression in tumor [100,106] and Smad7 overexpression have been assessed for their effect on tumor resistance and increasing the tumorigenicity in tumor cell lines [107]. Direct inhibition of Smad2/3 signaling has also been attempted but is difficult to be addressable with therapeutics. Only a few repurposed drugs (Halofuginone, specific Smad3 inhibitor, SIS3) have been used in clinical settings so far [104]. Since EMT is also involved in drug resistance in NSCLC, targeting EMT by combining pirfenidone with anti-cancer drug may help in ameliorating drug resistance [108-110]. Previously pirfenidone has been suggested to play a biphasic role in inhibition of EMT in non-small cell lung cancer [22]. However several of its molecular mechanisms remain to be elaborated [26]. In the present study we demonstrate the effect of chemotherapeutic agent, bleomycin on TGF-β/Smad signaling pathway and epithelial-to-mesenchymal transition in A-549 cells. This is evidenced by reduction in E-cadherin and upregulation of Vimentin. An early upregulation of TGF and rSmads-2,3,4,5 and iSmads-6,7 occur. In late phase, the iSmad-1 upregulates while Smads-6 and 7 downregulate leading to progression of EMT. We demonstrate that pirfenidone significantly inhibited EMT by inhibition of TGF-β1, Smad-2,3,4,5 expression and upregulating Smad-6,7. Pirfenidone reverses the EMT as evidenced by reduction in Vimentin and upregulation of E-cadherin expression and increases cell migration and invasive potential. This may help prevent the tumor cell metastasis. Pirfenidone could attenuate the EMT process induced not only by exogenous TGF-1 but also by paracrine TGF- produced from NSCLC cells. Further the decreased TGF-β production in NSCLC cells after pirfenidone treatment may attenuate chemoresistance of tumor cells [22]. Therefore, Pirfenidone is a promising new therapeutic agent for the treatment of NSCLC through the regulation of EMT [38]. This suggests the potential of repurposing the anti-fibrotic drug pirfenidone as adjuvant therapy for preventing progression of lung cancer. Thus emphasizing the utility of pirfenidone as a new adjuvant therapeutic agent for NSCLC patients.
5. Conclusions
In present study we demonstrate the efficacy of pirfenidone in reversing epithelial-mesenchymal transition (EMT) of A-549 cells in vitro by differentially regulating the TGF-β1 mediated Smad1-7 signaling pathway and modulating both E-Cadherin and Vimentin expression. The efficacy of pirfenidone in attenuating EMT in adenocarcinoma cells can be repurposed to reduced their migratory and invasive potential and reduce metastasis of non-small cell lung carcinoma cells.
Declarations
- Ethics approval and consent to participate: Yes, I have the approval of Institutional ethics committee for my project No-VPCI/IAEC/2019/24 dated 4/7/2019.
- Consent for publication: Yes, I have the Consent for publication from all the co- authors.
- Availability of data and materials – Yes, all the raw data is available with me.
- Competing interests – I hereby declare that there is no competing interest among the authors.
- Funding– ICMR -34/1/2019-TF/Nano/BMS, date: 20/5/2019.
- Authors' contributions-RK- conceptualized and performed experiments and interpreted the data and wrote manuscript. PK, AS, DSN, MR, HD, AP- Performed experiments and compiled data. MAK, DAK- reviewed manuscript and guided research at every step.
- Acknowledgements- I gratefully acknowledge, the grant given by ICMR -34/1/2019-TF/Nano/BMS, date: 20/5/2019 for carrying out the above research work.
References
- Campos-Balea B, de Castro Carpeño J, Massutí B, et al. Prognostic factors for survival in patients with metastatic lung adenocarcinoma: An analysis of the SEER database. Thorac Cancer 11(2020): 3357-3364.
- Popper HH. Progression and metastasis of lung cancer. Cancer and Metastasis Reviews 35 (2016): 75-91.
- Shin YS, Choi CH, Kim YJ, et al. Primary squamous cell carcinoma in the chest wall mimicking abscess. J Thorac Dis 7 (2015): E179.
- Tamura T, Kurishima K, Nakazawa K, et al. Specific organ metastases and survival in metastatic non?small?cell lung cancer. Mol Clin Oncol 3 (2015): 217-221.
- Riihimäki M, Hemminki A, Fallah M, et al. Metastatic sites and survival in lung cancer. Lung cancer 86 (2014): 78-84.
- Hendriks LE, Smit EF, Vosse BA, et al. EGFR mutated non-small cell lung cancer patients: more prone to development of bone and brain metastases?. Lung cancer 84 (2014): 86-91.
- Wilbertz T, Wagner P, Petersen K, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol 24 (2011): 944-953.
- Byers LA, Diao L, Wang J, et al. An epithelial–mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res 19 (2013): 279-290.
- Ahmadi A, Najafi M, Farhood B, et al. Transforming growth factor-β signaling: Tumorigenesis and targeting for cancer therapy. J Cell Physiol 234 (2019): 12173-12187.
- Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci 20 (2019): 2767.
- Friese MA, Wischhusen J, Wick W, et al. RNA interference targeting transforming growth factor-β enhances NKG2D-mediated antiglioma immune response, inhibits glioma cell migration and invasiveness, and abrogates tumorigenicity in vivo. Can. Res 64 (2004): 7596-7603.
- Pang L, Li Q, Wei C, et al. TGF-β1/Smad signaling pathway regulates epithelial-to-mesenchymal transition in esophageal squamous cell carcinoma: in vitro and clinical analyses of cell lines and nomadic Kazakh patients from northwest Xinjiang, China. PloS one 9 (2014): e112300.
- Derynck R, Zhang Y, Feng XH. Transcriptional activators of TGF-β responses: Smads.Cell95 (1998): 737-740.
- Yan X, Chen YG. Smad7: not only a regulator, but also a cross-talk mediator of TGF-β signalling.J 434 (2011): 1-10.
- Grattendick KJ, Nakashima JM, Feng L, et al. Effects of three anti-TNF-α drugs: etanercept, infliximab and pirfenidone on release of TNF-α in medium and TNF-α associated with the cell in vitro. Int. Immunopharmacol 8 (2008): 679-687.
- Jin J, Togo S, Kadoya K, et al.Pirfenidone attenuates lung fibrotic fibroblast responses to transforming growth factor-β1.Respir Res20 (2019):
- Ruwanpura SM, Thomas BJ, Bardin PG. Pirfenidone: molecular mechanisms and potential clinical applications in lung disease.J. Respir. Cell Mol. Biol62 (2020): 413-422.
- Wang G, Zhou X, Guo Z, et al. The Anti-fibrosis drug Pirfenidone modifies the immunosuppressive tumor microenvironment and prevents the progression of renal cell carcinoma by inhibiting tumor autocrine TGF-β. Cancer Biol Ther 23 (2022): 150-162.
- Burghardt I, Tritschler F, Opitz CA, et al. Pirfenidone inhibits TGF-β expression in malignant glioma cells. Biochem. Biophys. Res. Commun 354 (2007): 542-547.
- Kozono S, Ohuchida K, Eguchi D, et al. Pirfenidone inhibits pancreatic cancer desmoplasia by regulating stellate cells. Cancer Res 73 (2013): 2345-2356.
- Paliogiannis P, Fois SS, Fois AG, et al. Repurposing Anticancer Drugs for the Treatment of Idiopathic Pulmonary Fibrosis and Antifibrotic Drugs for the Treatment of Cancer: State of the Art. Curr Med Chem 28 (2021): 2234-2247.
- Fujiwara A, Shintani Y, Funaki S, et al. Pirfenidone plays a biphasic role in inhibition of epithelial-mesenchymal transition in non-small cell lung cancer.Lung Cancer106 (2017): 8-16.
- Yamamoto Y, Yano Y, Kuge T, et al. Safety and effectiveness of pirfenidone combined with carboplatin-based chemotherapy in patients with idiopathic pulmonary fibrosis and non-small cell lung cancer: A retrospective cohort study. Thoracic Cancer 11 (2020): 3317-3325.
- Hisatomi K, Mukae H, Sakamoto N, et al. Pirfenidone inhibits TGF-β1-induced over-expression of collagen type I and heat shock protein 47 in A549 cells.BMC Pulm. Med12 (2012): 1-9.
- Parvathaneni V, Kulkarni NS, Shukla SK, et al. Systematic Development and Optimization of Inhalable Pirfenidone Liposomes for Non-Small Cell Lung Cancer Treatment. Pharmaceutics 12 (2020): 206.
- Molina-Molina M, Machahua-Huamani C, Vicens-Zygmunt V, et al. Anti-fibrotic effects of pirfenidone and rapamycin in primary IPF fibroblasts and human alveolar epithelial cells.BMC Pulm. Med18 (2018): 1-13.
- Pijuan J, Barceló C, Moreno DF, et al. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol 7 (2019): 107.
- Aman A, Piotrowski T. Cell migration during morphogenesis.Biol 341 (2010): 20-33.
- Ramachandran A, Vizán P, Das D, et al. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition.Elife7 (2018): e31756.
- Xie M, He C, Wei S. Relationship between Expression of TGF-β1, Smad2, Smad4 and Prognosis of Patients with Resected Non-small Cell Lung Cancer.J. Cancer18 (2015): 543-548.
- Zhang S, Che D, Yang F, et al. Tumor-associated macrophages promote tumor metastasis via the TGF-β/SOX9 axis in non-small cell lung cancer.Oncotarget8 (2017): 99801.
- Roberts AB, Anzano MA, Lamb LC, et al. New class of transforming growth factors potentiated by epidermal growth factor: isolation from non-neoplastic tissues.Natl. Acad. Sci78 (1981): 5339-5343.
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells.Nature554 (2018): 544-548.
- Vázquez PF, Carlini MJ, Daroqui MC, et al. TGF-beta specifically enhances the metastatic attributes of murine lung adenocarcinoma: implications for human non-small cell lung cancer.Clin?. Exp. Metastasis30 (2013): 993-1007.
- Hu HH, Chen DQ, Wang YN, et al. New insights into TGF-β/Smad signaling in tissue fibrosis.Biol. Interact292 (2018): 76-83.
- Pickup MW, Owens P, Moses HL. TGF-β, bone morphogenetic protein, and activin signaling and the tumor microenvironment.Cold Spring Harb. Perspect. Biol9 (2017): a022285.
- Li J, Shen C, Wang X, et al. Prognostic value of TGF-β in lung cancer: systematic review and meta-analysis.BMC cancer19 (2019): 1-9.
- Fujiwara A, Funaki S, Fukui E, et al. Effects of pirfenidone targeting the tumor microenvironment and tumor-stroma interaction as a novel treatment for non-small cell lung cancer.Rep10 (2020): 1-13.
- TodaM,MizuguchiS,MinamiyamaY,et al. Pirfenidone suppresses polarization to M2 phenotype macrophages and the fibrogenic activity of rat lung fibroblasts.J Clin Biochem Nutr63 (2018): 58-65.
- Oku H, Shimizu T, Kawabata T, et al. Antifibrotic action of pirfenidone and prednisolone: different effects on pulmonary cytokines and growth factors in bleomycin-induced murine pulmonary fibrosis.J. Pharmacol590 (2008): 400-408.
- Macías-Barragán J, Sandoval-Rodríguez A, Navarro-Partida J, et al. The multifaceted role of pirfenidone and its novel targets.Fibrogenesis tissue repair3 (2010): 1-11.
- Choi K, Lee K, Ryu SW, et al. Pirfenidone inhibits transforming growth factor-β1-induced fibrogenesis by blocking nuclear translocation of Smads in human retinal pigment epithelial cell line ARPE-19.Vis18 (2012): 1010.
- Qin W, Liu B, Yi M, et al. Antifibrotic agent pirfenidone protects against development of radiation-induced pulmonary fibrosis in a murine model.Res190 (2018): 396-403.
- Lv Q, Wang J, Xu C, et al. Pirfenidone alleviates pulmonary fibrosis in vitro and in vivo through regulating Wnt/GSK-3β/β-catenin and TGF-β1/Smad2/3 signaling pathways. Mol. Med 26 (2020): 1-10.
- Bhowmick NA, Chytil A, Plieth D, et al. TGF-ß signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia.Science 303 (2004): 848-851.
- Kalluri R, Zeisberg M. Fibroblasts in cancer.Rev. Cancer ?6 (2006): 392-401.
- Yamada S, Nakamura J, Asada M, et al. Twisted gastrulation, a BMP antagonist, exacerbates podocyte injury.PloS one9 (2014): e89135.
- Munoz-Felix JM, Gonzalez-Nunez M, Martinez-Salgado C, et al. TGF-β/BMP proteins as therapeutic targets in renal fibrosis. Where have we arrived after 25 years of trials and tribulations?.Ther156 (2015): 44-58.
- Zhou C, Zeldin Y, Baratz ME,et al.Investigating the effects of Pirfenidone on TGF-β1 stimulated non-SMAD signaling pathways in Dupuytren’s disease -derived fibroblasts.BMC Musculoskelet Disord135 (2019).
- Ballester B, Milara J, Cortijo J. Pirfenidone anti-fibrotic effects are partially mediated by the inhibition of MUC1 bioactivation. Oncotarget 11 (2020): 1306.
- Wnuk D, Paw M, Ryczek K, et al. Enhanced asthma-related fibroblast to myofibroblast transition is the result of profibrotic TGF-β/Smad2/3 pathway intensification and antifibrotic TGF-β/Smad1/5/(8)9 pathway impairment. Sci Rep 10 (2020): 16492.
- Zeisberg M, Hanai JI, Sugimoto H, et al. BMP-7 counteracts TGF-β1–induced epithelial-to-mesenchymal transition and reverses chronic renal injury.Med9 (2003): 964-968.
- Chitra P, Saiprasad G, Manikandan R, et al. Berberine attenuates bleomycin induced pulmonary toxicity and fibrosis via suppressing NF-κB dependant TGF-β activation: a biphasic experimental study.Lett219 (2013): 178-193.
- Chen F, Weinberg RA. Biochemical evidence for the autophosphorylation and transphosphorylation of transforming growth factor beta receptor kinases.Natl. Acad. Sci92 (1995): 1565-1569.
- Liu B, Rong Y, Sun D, et al. Costunolide inhibits pulmonary fibrosis via regulating NF-kB and TGF-β1/Smad2/Nrf2-NOX4 signaling pathways.Biophys. Res. Commun510 (2019): 329-333.
- Yang YC, Piek E, Zavadil J, et al. Hierarchical model of gene regulation by transforming growth factor β.Natl. Acad. Sci100 (2003): 10269-10274.
- Higashiyama H, Yoshimoto D, Okamoto Y, et al. Receptor-activated Smad localisation in Bleomycin-induced pulmonary fibrosis. Journal of clinical pathology 60 (2007): 283-289.
- Flanders K, Sato M, Ooshima A, et al. Smad?3 as a mediator of the fibrotic response.J. Clin. Exp. Pathol 85 (2004): A13-A13.
- Piek E, Ju WJ, Heyer J, et al. Functional characterization of transforming growth factor β signaling in Smad2-and Smad3-deficient fibroblasts.Biol. Chem276 (2001): 19945-19953.
- Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-β/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach.Biol. Chem276 (2001): 17058-17062.
- Organ LA, Duggan AM, Oballa E, et al. Leeming DJ. Biomarkers of collagen synthesis predict progression in the PROFILE idiopathic pulmonary fibrosis cohort. Respiratory research 20 (2019): 1-0.
- Meng F, Li J, Yang X, et al. Role of Smad3 signaling in the epithelial?mesenchymal transition of the lens epithelium following injury. International journal of molecular medicine 42 (2018): 851-860.
- Shi N, Wang Z, Zhu H, et al. Research progress on drugs targeting the TGF-β signaling pathway in fibrotic diseases. Immunol Res (2022).
- Das S, Kumar M, Negi V, et al. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am J Respir Cell Mol Biol 50 (2014): 882-892.
- Aschenbrenner JK, Sollinger HW, Becker BN, et al. 1, 25-(OH2) D3 alters the transforming growth factor β signaling pathway in renal tissue.Surg. Res 100 (2001): 171-175.
- Monteleone G, Kumberova A, Croft NM, et al. Blocking Smad7 restores TGF-β1 signaling in chronic inflammatory bowel disease.Clin. Investig108 (2001): 601-609.
- Du X, Pan Z, Li Q, et al. SMAD4 feedback regulates the canonical TGF-β signaling pathway to control granulosa cell apoptosis.Cell Death Dis92 (2018): 1-12.
- Huang Y, He Y, Li J. MicroRNA-21: a central regulator of fibrotic diseases via various targets.Pharm. Des21 (2015): 2236-2242.
- Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol14 (2018): 111.
- Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders.Cell103 (2000): 295-309.
- Parveen A, Akash MSH, Rehman K, et al. Dual role of p21 in the progression of cancer and its treatment.Rev. Eukaryot. Gene Expr261 (2016): 49-62.
- Song HW, Kumar BK, Kim SH, et al. Agmatine Enhances Neurogenesis by Increasing ERK1/2 Expression, and Suppresses Astrogenesis by Decreasing BMP 2,4 and SMAD 1,5,8 Expression in Subventricular Zone Neural Stem Cells. Life Sci 89 (2011): 439-449.
- Meng XM, Chung AC, Lan HY. Role of the TGF-β/BMP-7/Smad pathways in renal diseases.Sci124 (2013): 243-254.
- Dudas PL, Argentieri RL, Farrell FX. BMP-7 fails to attenuate TGF-β1-induced epithelial-to-mesenchymal transition in human proximal tubule epithelial cells.Dial. Transplant24 (2009): 1406-1416.
- Macias MJ, Martin-Malpartida P, Massagué J. Structural determinants of Smad function in TGF-β signaling.Trends Biochem40 (2015): 296-308.
- De Langhe E, Cailotto F, De Vooght V, et al. Enhanced endogenous bone morphogenetic protein signaling protects against bleomycin induced pulmonary fibrosis.Res16 (2015): 1-10.
- Ming-Li Z, Zhong-Hua C, Ying-Ying T, et al. The Smad Dependent TGF-β and BMP Signaling Pathway in Bone Remodeling and Therapies. Frontiers in Mol Biosci 8 (2021).
- Kim MS, Jin W. TrkB-Induced Inhibition of R-SMAD/SMAD4 Activation Is Essential for TGF-β-Mediated Tumor Suppressor Activity. Cancers 12 (2020): 1048.
- Zhao J, Shi W, Chen H, et al. Smad7 and Smad6 differentially modulate transforming growth factor β-induced inhibition of embryonic lung morphogenesis.Biol. Chem275 (2000): 23992-23997.
- Mami S, Ghaffarpour S, Faghihzadeh S, et al. Evaluation of the LTBP1 and Smad6 Genes Expression in Lung Tissue of Sulfur Mustard-exposed Individuals with Long-term Pulmonary Complications.Iran J Allergy Asthma Immunol (2019): 473-478.
- Isaka Y. Targeting TGF-β signaling in kidney fibrosis. International journal of molecular sciences19 (2018): 2532.
- Sharma K, Ix JH, Mathew AV, et al. Pirfenidone for diabetic nephropathy. Journal of the American Society of Nephrology 22 (2011): 1144-1151.
- Nakao A, Fujii M, Matsumura R, et al. Transient gene transfer and expression of Smad7 prevents bleomycin-induced lung fibrosis in mice.Clin. Investig104 (1999): 5-11.
- Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses.Cell25 (2005): 8108-8125.
- Dawes LJ, Sleeman MA, Anderson IK, et al. TGFβ/Smad4-dependent and-independent regulation of human lens epithelial cells.Ophthalmol. Vis. Sci50 (2009): 5318-5327.
- Chen Y, Zhang Q, Zhou Y, et al. Inhibition of miR-182-5p attenuates pulmonary fibrosis via TGF-β/Smad pathway. Hum. Exp. Toxicol 39 (2020): 683-695.
- Bi L, Huang Y, Li J, et al. Pirfenidone Attenuates Renal Tubulointerstitial Fibrosis through inhibiting miR-21. Nephron 146 (2022): 110-120.
- Weng CM, Li Q, Chen KJ, et al. Bleomycin induces epithelial-to-mesenchymal transition via bFGF/PI3K/ESRP1 signaling in pulmonary fibrosis. Biosci Rep 40 (2020): BSR20190756.
- Agiostratidou G, Hulit J, Phillips GR, et al. Differential cadherin expression: potential markers for epithelial to mesenchymal transformation during tumor progression. J Mammary Gland Biol Neoplasia 12 (2007): 127-133.
- Aban CE, Lombardi A, Neiman G, et al. Downregulation of E-cadherin in pluripotent stem cells triggers partial EMT.Rep11 (2021): 1-11.
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition.Rev. Mol. Cell Biol15 (2014): 178-196.
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease.Cell139 (2009): 871-890.
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 331 (2011): 1559-1564.
- Chen KJ, Li Q, Wen CM, et al. Bleomycin (BLM) induces epithelial-to-mesenchymal transition in cultured A549 cells via the TGF-β/Smad signaling pathway.Cancer7 (2016): 1557.
- Kurimoto R, Ebata T, Iwasawa S, et al. Pirfenidone may revert the epithelial-to-mesenchymal transition in human lung adenocarcinoma.Lett14 (2017): 944-950.
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition.Clin. Investig119 (2009): 1420-1428.
- Guo J, Yang Z, Jia Q, et al. Pirfenidone inhibits epithelial-mesenchymal transition and pulmonary fibrosis in the rat silicosis model. Toxicology letters 300 (2019): 59-66.
- Shintani Y, Fujiwara A, et al. IL-6 secreted from cancer-associated fibroblasts mediates chemoresistance in NSCLC by increasing epithelial-mesenchymal transition signaling.J Thorac Oncol11 (2016): 1482-1492.
- Katz LH, Likhter M, Jogunoori W, et al. TGF-β signaling in liver and gastrointestinal cancers.Cancer Lett379 (2016): 166-172.
- Shany S, Sigal-Batikoff I, Lamprecht S. Vitamin D and myofibroblasts in fibrosis and cancer: At cross-purposes with TGF-β/SMAD signaling.Anticancer res36 (2016): 6225-6234.
- Jung B, Staudacher JJ, Beauchamp D. Transforming growth factor β superfamily signaling in development of colorectal cancer.Gastroenterology152 (2017): 36-52.
- Eser PÖ, Jänne PA. TGFβ pathway inhibition in the treatment of non-small cell lung cancer.Ther184 (2018): 112-130.
- Zeng Y, Zhu J, Shen D, et al. MicroRNA-205 targets SMAD4 in non-small cell lung cancer and promotes lung cancer cell growth in vitro and in vivo.Oncotarget8 (2017): 30817.
- Trendelenburg AU. TGFβ Signaling (2020): 1-36.
- Thiery JP. Epithelial–mesenchymal transitions in tumour progression.Rev. Cancer?2 (2002): 442-454.
- Chaikuad A, Bullock AN. Structural basis of intracellular TGF-β signaling: receptors and smads.Cold Spring Harb. Perspect. Biol8 (2016): a022111.
- Luo L, Li N, Lv N, et al. SMAD7: a timer of tumor progression targeting TGF-β signaling.Tumor Biol35 (2014): 8379-8385.
- Shintani Y, Okimura A, Sato K, et al. Epithelial to mesenchymal transition is a determinant of sensitivity to chemoradiotherapy in non-small cell lung cancer.Thorac. Surg92 (2011): 1794-1804.
- Li S, Xu A, Li Y, et al. RS4651 suppresses lung fibroblast activation via the TGF-β1/SMAD signalling pathway. European Journal of Pharmacology 903 (2021): 174135.
- Zeng Z, Qing YC, Hu Z, et al. Distinct expression and prognostic value of members of SMAD family in non-small cell lung cancer, Medicine 99 (2020):