Next Generation Sequencing Approach and Impact on Bioinformatics: Applications in Agri-Food Field
Article Information
Tiziana Maria Sirangelo1*, Grazia Calabrò2
1Tiziana Maria Sirangelo Life Science Department, University of Modena and Reggio Emilia, Italy
2Computer Engineering Department. University of Calabria, Italy
*Corresponding Author: Tiziana Maria Sirangelo, Tiziana Maria Sirangelo Life Science Department, University of Modena and Reggio Emilia, Italy
Received: 04 April 2020; Accepted: 16 April 2020; Published: 20 April 2020
Citation: Tiziana Maria Sirangelo, Grazia Calabrò. Next Generation Sequencing Approach and Impact Oon Bioinformatics: Applications in Agri-Food Field. Journal of Bioinformatics and Systems Biology 3 (2020): 032-044.
Share at FacebookAbstract
The advances in omics fields and the growth of high-throughput biological data required the use of bioinformatics tools to elaborate complex data, as well as the development of network resources, such as databases referred to specific biological fields and shared by the scientific community. This work mentions some of the main typologies of software tools used in the standard workflow of the NGS approach, focusing on the main problems associated with their use. Crucial applications in agri-food fields are also described. Particularly, in agricultural area it is highlighted how advanced bioinformatic tools made genome sequencing of key species possible, such as rice, grapevine, corn, tomato, potato, peach, barley, while numerous other sequencing projects are nearing completion. It is also shown how metagenomics approaches and complex software packages were applied to soil microbiome investigation. In the food microbiology area it is underlined how today the metagenomic techniques based on amplicon sequencing as well as on whole genome sequencing are widely used for analysing the microbial composition of products and for studying the microbiome and the mycobiome in the fermentative processes. Finally, some current needs are discussed. Among them relevant ones are those related to the elaboration of big amount of data provided by NGS data analysis, requiring new and more powerful bioinformatics resolutions to handle such large biological collections. Furthermore, specialized software tools and advanced computational resources for data integration are necessary.
Keywords
Next Generation Sequencing; Bioinformatics; Agri-food field
Next Generation Sequencing articles, Bioinformatics articles, Agri-food field articles
Next Generation Sequencing articles Next Generation Sequencing Research articles Next Generation Sequencing review articles Next Generation Sequencing PubMed articles Next Generation Sequencing PubMed Central articles Next Generation Sequencing 2023 articles Next Generation Sequencing 2024 articles Next Generation Sequencing Scopus articles Next Generation Sequencing impact factor journals Next Generation Sequencing Scopus journals Next Generation Sequencing PubMed journals Next Generation Sequencing medical journals Next Generation Sequencing free journals Next Generation Sequencing best journals Next Generation Sequencing top journals Next Generation Sequencing free medical journals Next Generation Sequencing famous journals Next Generation Sequencing Google Scholar indexed journals Agri-food field articles Agri-food field Research articles Agri-food field review articles Agri-food field PubMed articles Agri-food field PubMed Central articles Agri-food field 2023 articles Agri-food field 2024 articles Agri-food field Scopus articles Agri-food field impact factor journals Agri-food field Scopus journals Agri-food field PubMed journals Agri-food field medical journals Agri-food field free journals Agri-food field best journals Agri-food field top journals Agri-food field free medical journals Agri-food field famous journals Agri-food field Google Scholar indexed journals DNA/RNA molecules articles DNA/RNA molecules Research articles DNA/RNA molecules review articles DNA/RNA molecules PubMed articles DNA/RNA molecules PubMed Central articles DNA/RNA molecules 2023 articles DNA/RNA molecules 2024 articles DNA/RNA molecules Scopus articles DNA/RNA molecules impact factor journals DNA/RNA molecules Scopus journals DNA/RNA molecules PubMed journals DNA/RNA molecules medical journals DNA/RNA molecules free journals DNA/RNA molecules best journals DNA/RNA molecules top journals DNA/RNA molecules free medical journals DNA/RNA molecules famous journals DNA/RNA molecules Google Scholar indexed journals biological data articles biological data Research articles biological data review articles biological data PubMed articles biological data PubMed Central articles biological data 2023 articles biological data 2024 articles biological data Scopus articles biological data impact factor journals biological data Scopus journals biological data PubMed journals biological data medical journals biological data free journals biological data best journals biological data top journals biological data free medical journals biological data famous journals biological data Google Scholar indexed journals transcriptomic articles transcriptomic Research articles transcriptomic review articles transcriptomic PubMed articles transcriptomic PubMed Central articles transcriptomic 2023 articles transcriptomic 2024 articles transcriptomic Scopus articles transcriptomic impact factor journals transcriptomic Scopus journals transcriptomic PubMed journals transcriptomic medical journals transcriptomic free journals transcriptomic best journals transcriptomic top journals transcriptomic free medical journals transcriptomic famous journals transcriptomic Google Scholar indexed journals genomic articles genomic Research articles genomic review articles genomic PubMed articles genomic PubMed Central articles genomic 2023 articles genomic 2024 articles genomic Scopus articles genomic impact factor journals genomic Scopus journals genomic PubMed journals genomic medical journals genomic free journals genomic best journals genomic top journals genomic free medical journals genomic famous journals genomic Google Scholar indexed journals metagenomics articles metagenomics Research articles metagenomics review articles metagenomics PubMed articles metagenomics PubMed Central articles metagenomics 2023 articles metagenomics 2024 articles metagenomics Scopus articles metagenomics impact factor journals metagenomics Scopus journals metagenomics PubMed journals metagenomics medical journals metagenomics free journals metagenomics best journals metagenomics top journals metagenomics free medical journals metagenomics famous journals metagenomics Google Scholar indexed journals transcriptomics articles transcriptomics Research articles transcriptomics review articles transcriptomics PubMed articles transcriptomics PubMed Central articles transcriptomics 2023 articles transcriptomics 2024 articles transcriptomics Scopus articles transcriptomics impact factor journals transcriptomics Scopus journals transcriptomics PubMed journals transcriptomics medical journals transcriptomics free journals transcriptomics best journals transcriptomics top journals transcriptomics free medical journals transcriptomics famous journals transcriptomics Google Scholar indexed journals De novo genomic articles De novo genomic Research articles De novo genomic review articles De novo genomic PubMed articles De novo genomic PubMed Central articles De novo genomic 2023 articles De novo genomic 2024 articles De novo genomic Scopus articles De novo genomic impact factor journals De novo genomic Scopus journals De novo genomic PubMed journals De novo genomic medical journals De novo genomic free journals De novo genomic best journals De novo genomic top journals De novo genomic free medical journals De novo genomic famous journals De novo genomic Google Scholar indexed journals
Article Details
Introduction
Next Generation Sequencing (NGS) technologies, which offer high-throughput methods to investigate sequences of nucleotides within DNA/RNA molecules [1], have now become an essential tool in the applications of the biological sciences. Bioinformatics is fundamental to the interpretation of these type of data, in fact, mathematical and statistical methods implemented by using several programming paradigms and dedicated software tools are able to analyse and explain biological molecular, cellular or genomic information This interdisciplinary field significantly started growing since the mid-1990s, thanks to the Human Genome Project and the fast progress in DNA sequencing technology. It played and still plays today a main role in research projects about crucial topics, like sequence alignment, gene finding, genome assembly and prediction of gene expression. Particularly, bioinformatics, in its early years was mainly about biological data management and their broadcast on Internet. Then, it was proved to be suitable for data analysis and modelling, making it possible not only to detect added value information from NGS data, but also to support the prediction of relevant properties from large and often heterogeneous data sets [2]. The adoption of NGS technologies highlighted the need for bioinformatics to process and interpret the massive amount of data coming from the sequencing tasks. Particularly, the advances in omics fields and the growth of high-throughput biological data required the use of bioinformatics resources and software tools to elaborate complex “Big Data”, otherwise unusable in practice. It was also necessary to develop network resources, such as databases referred to specific biological fields, structured data collections and vocabularies accessible and shared by the scientific community.
More in detail, referring to the application of a NGS-based approach, that can be summarized in three distinct steps, each of which addresses the transformation of raw data into a specific type of biological knowledge [3], many software tools are necessary to support it. Understanding the basic concepts of these steps is important in assessing the informatics needs of a given omics research project. The first two steps consist of processing raw sequencing signals into nucleotide bases and reads, and in the alignment of these reads to reference sequences, respectively. In the third step, the obtained genomic/transcriptomic profile is associated to a descriptive annotation, in order to give a biological interpretation to the results.
More specifically, the first step consists of a primary analysis in which the software tools are strictly interfaced with sequencing equipment, making it possible to convert the raw signal into nucleotides sequences. These tools are almost always installed on the hardware being part of the sequencing equipment. However, they can also be located separately, and may often be appropriate to improve the initial signal processing and consequently the overall data quality. Raw data are generally produced by sequencing centres, in form of fragmented sequences, to be pre-processed, i.e., by removing contaminations, or other specific sequences such as adaptors, barcodes, according to the specific technology used.
In the secondary analysis, an assembly step is necessary in order to reconstruct as accurately as possible the original sequence, on the basis of the results of the sequence alignment process. This procedure can use a guided approach if the alignment is performed by comparing already available reference sequences or a de novo approach if this comparison does not take place. Comparison with reference genomes/transcriptomes databases or other tailored data collections is very important in order to transfer information from already annotated molecules to others yet to be defined. This process also helps to detect peculiarities and provides hints for deeper investigations [4]. Once reads have been aligned to the reference genome/transcriptome, next steps are foreseen in order to filter duplicate reads, to realign and to minimize erroneous alignments.
The analyses performed in the third step support different kinds of investigations such as those required for a better structure definition, feature identification and taxonomic assignment.
Many bioinformatic tools and pipelines were developed in order to support the second and third step, as well as assembly tools, reference databases, annotations tools, genome browsers for data visualization, some of them are considered in the next section.
Bioinformatics tools in NGS approach
The standard workflow associated to the application of the NGS approach, related to the analysis of the omics data, is schematized in Fig 1. Data coming from biological samples (plant, soil, food) are sequenced by using a given omics kind of study (based on metagenomics, transcriptomics or genomics) and then processed by assembly or directly exposed to the prediction analyses carried out by using reference databases. The third step includes different investigations such as feature identification and taxonomic assignment.
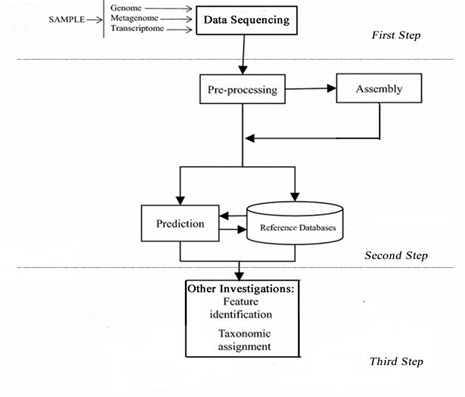
Figure 1: Standard workflow of omics data analysis
Some of the most common and used open-source software and reference databases in genomic, transcriptomic, and metagenomic studies are described below, making some relevant considerations from time to time and distinguishing them for each workflow phase.
Gene Identification and Sequence Analyses phase:
Sequence analyses refer to a proper understanding of different nucleic acid features and today are one of the most frequent applications of bioinformatics. Tools used for primary sequence analyses are widely described in scientific literature [5].
Reads pre-processing phase:
Among the most used software, it is possible to find:
- FastQC, for the quality check and report of NGS data [6]
- FASTX-toolkit, a package for the manipulation of sequence data and their format conversion [7].
Assembly and Alignment phase:
One of the most important steps in NGS analysis is de novo genome assembly and today advanced algorithms allow the assembly of complex eukaryotic genomes [8]. The assembly process is quite complicated and the understanding of the underlying methods is necessary in order to generate consistent and quality results. These algorithms are not described here also because there is a good number of investigations treating them [9].
In order to perform an accurate alignment of a big amount of sequences to a reference, many tools were implemented. The general principle behind these software packages is trying to detect the possible alignment locations and then actually carrying out the alignment.
Below, some of the most used and traditional assembly and alignment algorithms are listed:
- (META) VELVET/OASES, a De novo genomic/transcriptomic assembler [10]
- SOAP DE NOVO, a De novo short-read assembler [11]
- TRINITY, for a De novo assembly of RNA-seq data [12]
- BLAT (a BLAST-like alignment tool), designed for the alignment of sequences to a reference genome [13].
- Bowtie, a programme for the alignment of short nucleotide sequences to genomic sequence [14]
- Star, a RNA-seq to genome aligner [15]
- Tophat/Cufflinks, a RNA-seq to genome aligner and quantification tools [16].
Gene prediction/annotation phase:
High quality annotations optimize the effectiveness of using previously sequenced genomes. In fact, they make it possible to link genomic sequences to their biological function. Below, some of the most used and traditional annotation tools are shown:
- Ensembl genome annotation, a Gene annotation pipeline [17]
- Genemark, a Gene prediction software including unsupervised and semi-supervised training [18]
- NCBI genome annotation, a Genome annotation pipeline released by NCBI (National Centre for Biotechnology Information) [19].
Visualization tools, such as Genome Browsers, were developed, in order to integrate genomic sequence and annotation data from different sources and to allow browsing, retrieving and analysis of these data sets. Among the main Genome Browsers, we mention Ensemble Genome Browser and NCBI's Genome Browser, associated with the aforementioned software.
Reference databases:
The very same revolution in DNA-sequencing approach that has greatly reduced the overall sequencing cost has also resulted into a large increase in the volume of data generated. Furthermore, long reads produced by Sanger sequencing were replaced by short reads or their pairs. Sequence data management was thus affected by these new features, requiring new bioinformatics approaches, even in terms of storing data, viewing and using them [20]. New and more complex databases providing customized interfaces to process and query sequence data became necessary.
Below, some of the most used and traditional databases are mentioned:
- RDP/Silva/Greengenes are repositories of ribosomal RNA genes, used for supporting taxonomic annotation [21]
- KEGG are integrated resources for functional annotation of genes [22]
- UNIPROT, a database of functional annotated protein sequences [23]
- GenBank, built by the NCBI, is a large collection of genome sequences of over 250,000 species. Data can be accessed through the NCBI’s retrieval system called Entrez. The collection includes coding and untranslated regions, promoters, terminators, exons, introns, repeat regions [24].
The workflow shown in the figure is still valid even when a metagenomic approach is adopted. Given the importance that metagenomic analysis has played in agri-food area, here we make some pertinent considerations. For carrying out this type of investigation, two methods can be adopted. Traditionally, microbiomes analyses were focused on profiling taxonomic abundance by using the amplicon sequencing of 16 rRNA genes approach. Instead, in the last few years, the shotgun metagenomics method, consisting in the sequencing of all sequences of the genome, proved to be predominant and more detailed than the previous approach [25]. Mothur [26] and QIIME [27] are among the most used software tools, based on 16S data clustering and classification. Instead, for supporting a shotgun metagenome analysis, Megan [28] and Metamos [29] pipelines are generally used.
NGS and bioinformatics in agricultural field
Omics sciences efforts have now spread to all fields including agricultural research area. These researches can be about a single species (a population), or multiple species (a community). In the first case, the functionality of organs, for instance roots and fruits, are investigated, in order to individuate their main characteristics and their behaviour with reference to stress resistance, diseases, senescence, and so on [32]. Instead, while adopting a metagenomic approach, the microbial community can include samples coming from soil, salty or not water, roots [33]. In both cases, a deep knowledge of underlying mechanisms, made possible thanks to these innovative approaches, allowed to highlight the functionality of biological systems, to trace molecular variability during their development, in different conditions, like those due to environmental changes [30] that is also known to influence gene expression [31].
Nucleic acid sequencing and NGS technologies have enormously contributed to the progress in these fields [34;32]. Transcriptome and proteome sequencing were fundamental to obtain a deeper knowledge of the genome and of the main functionalities of many species in the agricultural area [32]. Moreover, detailed information about genetic maps were very useful to obtain a better stamina and resistance of crops, increasing their productivity. Genomics researches also resulted into innovative solutions for their protection from diseases [35] and for a more sustainable agriculture, with an enormous impact on food industry.
Soil biological and chemical properties affect plant quality features [36] and most studies about this research topic are now using whole DNA extraction and NGS-based metagenomic methods, making it possible to individuate the complicate interactions within soil microbiome and plant rhizosphere [37]. Metagenomics recently allowed to study the role of soil microbial community in plant nutrition [38], the changes in soil and in rhizosphere microbiome due to fertilization [39] and to organic farming [40], as well as to investigate bio-products [41] contributing to improve plant growth and to protect their health.
As a consequence of the spreading of NGS approaches, bioinformatics has acquired a crucial role in agricultural research field. High-throughput technologies, supported by advanced bioinformatic tools, allowed the genome sequencing of key species, such as Arabidopsis thaliana (The Arabidopsis Genome Initiative 2000) and later of other relevant ones, such as rice, grapevine, corn, apple tree, tomato, potato, peach, barley. Meanwhile, numerous other sequencing projects are nearing completion [42]. These efforts were also accompanied by transcriptome sequencing research activities based on different bioinformatics technologies [43], often requiring appropriate software resources [44], as well as suitable pipelines able to move from raw to integrated biological data [45]. Specific big data collections, from transcriptomics and metagenomics projects using these new technologies, were created, and therefore dedicated storage system were used. Among them, a common one is the Sequence Read Archive (SRA) [46], which may be considered only a small part of the storage collections today available. Many bioinformatic efforts were also made in order to manage in optimal way these large datasets.
On the other hand, the increasing number of reference genomes addressed research efforts towards the genome variation study, in order to obtain a dataset of mutations from individual genomes, like Single Nucleotide Polymorphisms (SNPs). SNPs are the most abundant type of DNA sequence variation in plants, and, due the importance of use these large data collections for the species of agricultural interest, several bioinformatics tools were developed to allow their browsing [47].
NGS and bioinformatics in food microbiology field
In the last decades, food microbiology took advantage of the most recent advances in molecular biology and new techniques to detect and monitor microorganisms have been adopted [48]. This evolution went hand in hand with environmental microbiology, the field that first used them. More specifically, the introduction of high-throughput sequencing made possible more accurate and reliable studies of food microbial communities [49;50]. Compared to the traditional culture-dependent techniques, the number of nucleic acids sequenced with NGS techniques is far higher, making it easier to detail the composition of food bacterial population [51] and to monitor the changes that take place over time. Therefore, it is possible, for instance, to obtain more detailed information about the dynamics of fermentation processes and the growth of starter cultures [52]. As NGS technologies improved, the read length and the overall quality of the sequences have also improved, making possible to identify species with a greater resolution. Particularly, a study showed how adopting the NGS metagenomic approach to characterize raw milk microbiota can highlight the changes that occur in the different stages of production and storage conditions [53]. Being generally a highly suitable tool for characterizing the genetic potential of many types of microbiota, culture-independent metagenomic approaches have been successfully applied to several fields, including food area. An analysis of accuracy and speed of some analytical metagenomics tools is found in a paper [54].
Amplicon sequencing is one of the most popular metagenomic techniques for analysing the microbial composition of the foods, as well as studying the microbiome and mycobiome in the fermentative process [55]. On the other side, in recent years, the metagenomic based on a whole genome sequencing has been applied to food microbiology and significant results have been achieved, allowing researchers to obtain profiles of species yet to be characterized [56].
However, other omics investigations can also be applied to the food field: metatranscriptomics techniques make it possible to study the functional aspects of the food microbiota, whereas metaproteomic approaches and the identification of protein profiles can be used to create classes of marker compounds of many food properties [57]. Furthermore, metabolomic methods are used to identify and quantify metabolites in food, as well as to monitor changes during dynamic processes such as fermentation, and finally to study and hypothesize the metabolic pathway behind their production. The application of omics methods goes beyond the identification of the microorganisms present in foods and focuses on their role in a more complex network [58]. An integrated approach which includes a proper combination of data coming from each omics field is able to provide promising insights for characterizing dynamic processes as well as to monitor the effects of starter cultures on food evolution. More recently, NGS based techniques were adopted for similar studies, showing promising results. An example of that can be found in an article [59], in which culture dependent techniques and metagenomics approaches are used for the characterization of spoilage organisms in vacuum-packed cooked ham.
Consequently, bioinformatics is increasingly used in many applications of food microbiology, such as food fermentation and safety. Generally, it could be desirable to have databases using controlled vocabularies to integrate data from genomics, systems biology, phenotypes, and that are designed on the basis of the FAIR (findable, accessible, interoperable, re-usable) approach in storing and managing data [60]. Today, a large variety of databases storing data on food, their constituents and nutritive values are available [61]. Particularly, about food fermentation, innovative datasets related to genotype/phenotype/transcriptome such as those available for L. lactis and L. plantarum could help make new sequence-based functional prediction strategies to select, for instance, carbohydrate active enzymes [62]. In food safety area, projects are now focusing on the pathogens and innovative ways to detect the source of the food borne illnesses [63]. Both in food safety and fermentation, the function prediction from sequence data is fundamental and in order to reach this goal, bioinformatics plays a crucial role and only advanced algorithms can led to reliable results.
Conclusions
The high-throughput methods make it possible to massively output data very quickly, but such results are not immediately available for biological aims, and must necessarily be subjected to bioinformatics analysis.
Thus, bioinformatics tools work on datasets that are usually very large and consequently a strong calculation power and a large storage space are often required. The big amount of data provided by NGS data analysis needs new and more powerful bioinformatics solutions able to manage large biological collections, and also specialized software tools and advanced computational resources for their analysis and integration.
This is particularly true when such data come from a multi-omics integrated approach, that is now known to be powerful enough to elucidate the ecologic role of microbiomes in agri-food contexts [66].
In this case, a detailed data requirements analysis is very important, in order to develop a computational pipeline able to integrate data generated generally from different platforms. Therefore, it is clear that specific bioinformatic approaches are necessary to appropriately support the analysis of complex biological information and their integration, like that produced while merging transcriptomic, proteomics or metabolomics results.
In the agri-food field, at the moment, several tools are available to investigate the microbial community, but, according to many recent studies, a system level approach is still lacking [67]. Only few researches try to adopt this approach in multi-omics applications. Among them, an example is a research work about biological wastewater treatment [68], but also extensible to other areas. Its aim is to define a computational framework able to individuate a large microbial community by the integration of multi-omics data.
It is also necessary to have integrated software network resources, accessible and shared to the whole scientific community. Continuous concerning genome sequences updates, as well as the introduction of new annotations or RNAseq data sets, highlight the need for databases associated to different data collections, but at the same time “open” (constantly update and accessible) and as close to standard as possible [64]. The aim is to avoid, for instance, different gene annotations for the same genome as well as misalignments between data and the content of the used reference database.
Organizing, sharing and integrating biological data is contributing to spread new kinds of resources and common methods. This resulted into a revolution of agri-food practice and production processes, offering knowledge and tools to improve quality. Furthermore, agricultural strategies of protection against environmental stress, diseases, and parasites can be better designed and developed. The different applications based on this innovative scientific knowledge are also essential for providing novel products and applications for crop management.
Moreover, novel bioinformatics tools making it possible to integrate data from different software environments are also required, especially when used in the de novo assembly process [65]. In fact, even though several assembly tools have been implemented and customized for the reconstruction of genomes/transcriptomes from short reads, further efforts are necessary to improve the performance of assembly algorithms.
In the light of this, specific and tailored bioinformatics and biostatistics skills for NGS data analysis and for computational pipelines development are necessary, particularly when multi-omics approaches are implemented. At the same time, in order to make the best use of the obtained data, it is important to rely on bioinformatic analysts, able to appropriately use the available resources and, if necessary, to develop new software tools, with the aim to produce quality and standardized results. They must be able of using advanced software, algorithms, of managing databases and networking technologies, to analyse and explain high-throughput complex biological data, sharing vocabularies as well as research results.
References
- Metzker ML. Sequencing technologies: the next generation. Nat Rev Genet 11 (2010): 31–46.
- Chiusano ML, On the Multifaceted Aspects of Bioinformatics in the Next Generation Era: The Run that must keep the Quality. Next Generat Sequenc & Applic 2 (2015).
- Oliver GR, Hart SN, Klee EW. Bioinformatics for Clinical Next Generation Sequencing. Clinical Chemistry 61 (2015): 124–135.
- Mathé C, Sagot MF, Schiex T, Rouze P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res 30 (2002): 4103–17.
- Mehmood MA, Sehar U & Ahmad N. Use of bioinformatics tools in different spheres of life sciences. Data Mining Genomics Proteomics 5 (2014): 158.
- Wingett SW, Andrews S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 7 (2018): 1338.
- Patel RK, Jain M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 7 (2012): e30619.
- Edwards D, Batley J. Plant genome sequencing: applications for crop improvement, Plant Biotechnol J 7 (2010): 1-8.
- Zhang W, Chen J, Yang Y, et al. A practical comparison of de novo genome assembly software tools for next-generation sequencing technologies Plos One 6 (2011): e17915.
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 8 (2012): 1086–92.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1 (2012): 18.
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol 29 (2011): 644.
- Kent W. BLAT - the BLAST-like alignment tool, Genome Res 12 (2002): 656-64.
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol 10 (2009): R25.
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (2013): 15–21.
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7 (2012): 562–78.
- Aken BL, Ayling S et al., The Ensembl gene annotation system. Database: The Journal of Biological Databases and Curation (2016): baw093.
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33 (2005): 6494–506.
- Thibaud-Nissen F, Souvorov A, Murphy T, DiCuccio M, and Kitts P. Eukaryotic Genome Annotation Pipeline. The NCBI Handbook 2nd edition. (2013).
- Lee HC, Lai K, Tadeusz Lorenc M, Imelfort M, Duran C, Edwards D. Bioinformatics tools and databases for analysis of next-generation sequence data. Briefings in Functional Genomics 11 (2012):12–24.
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 3 (2012): 610–8.
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28 (2000): 27–30.
- The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research 47 (2019): D506–D515,
- Benson DA, Karsch-Mizrach I, Lipman DJ. Genbank. Nucleic Acids Research 30 (2002): 17-20.
- Shakya M, Lo C and Chain PSG. Advances and Challenges in Metatranscriptomic Analysis. Frontiers in Genetics 10 (2019). Article 904.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75 (2009): 7537–41.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7 (2010): 335–6.
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res 17 (2007): 377–86.
- Treangen TJ, Koren S, Sommer DD, Liu B, Astrovskaya I, Ondov B, Darling AE, Phillippy AM, Pop M. MetAMOS: a modular and open source metagenomic assembly and analysis pipeline. Genome Biol 14 (2013): R2.
- Barh D, Zambare V, Azevedo V, Omics. Applications in biomedical, agricultural, and environmental sciences. Boca Raton: CRC Press (2013).
- Ekblom R, Wolf JBW. A field guide to whole-genome sequencing, assembly and annotation. Evol Appl 7 (2014): 1026–42.
- Van Emon J. Omics revolution in agricultural research. J Agri Food Chem (2015).
- Meneghine AK, Nielsen S, Varani AM, Thomas T, Carareto Alves LM. Metagenomic analysis of soil and freshwater from zoo agricultural area with organic fertilization. PLoS ONE 12 (2017): e0190178.
- Barabaschi D, Tondelli A, Desiderio F, Volante A, Vaccino P, Valè G, Cattivelli L. Next generation breeding. Plant Sci (2015).
- Van der Vlugt R, Minafra A, Olmos A, Ravnikar M, Wetzel T, Varveri C, Massart S. Application of next generation sequencing for study and diagnosis of plant viral diseases in agriculture (2015).
- Acosta-Martínez V, Cotton J, Gardner T, Moore-Kucera J, Zak J, Wester D, Cox S. Predominant bacterial and fungal assemblages in agricultural soils during a record drought/heat wave and linkages to enzyme activities of biogeochemical cycling. Applied Soil Ecology 84 (2014): 69–82.
- Wang Z, Li T, Wen X, Liu Y, Han J, Liao Y and Jennifer M. DeBruyn Fungal.Communities in Rhizosphere Soil under Conservation Tillage Shift in Response to Plant Growth. Front. Microbiol 11 (2017).
- Pii Y, Borruso L, Brusetti L, Crecchio C, Cesco S, Mimmo T. The interaction between iron nutrition, plant species and soil type shapes the rhizosphere microbiome. Plant Physiol Biochem 99 (2016): 39–48.
- Pan Y, Cassman N, de Hollander M, Mendes LW, Korevaar H, Geerts RH, van Veen JA, Kuramae EE. Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol Ecol 90 (2014): 195–205.
- Bonanomi G, De Filippis F, Cesarano G, La Storia A, Ercolini D, Scala F. Organic farming induces changes in soil microbiota that affect agro-ecosystem functions. Soil Biology and Biochemistry 103 (2016): 327-336.
- Zhang Q, Acuña JJ, et al. Endophytic Bacterial Communities Associated with Roots and Leaves of Plants Growing in Chilean Extreme Environments. Sci Rep 9 (2019): 4950.
- Agarwal R & Narayan J. Unraveling the impact of bioinformatics and omics in agriculture. Int J Plant Biol Res 3 (2015): 1039.
- Iquebal M, Jaiswal S, Mukhopadhyay C, Sarkar C, Rai A, Kumar D: Applications of Bioinformatics in Plant and Agriculture in PlantOmics: The Omics of Plant Science. Springer (2015): 755-89.
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST–database for “expressed sequence tags”. Nat Genet 4 (1993): 332–3.
- Haas BJ, Papanicolaou A, Yassour M. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity, Nat Protoc 8 (2014).
- Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res (2010).
- Aflitos SA, Sanchez-Perez G, Ridder D, Fransz P, Schranz ME, Jong H, Peters SA. Introgression browser: high-throughput whole-genome SNP visualization. Plant J 82 (2015): 174–82.
- De Filippis F, La Storia A, Villani F, Ercolini D. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. Plos One 8 (2013): e70222.
- Bokulich NA, Lewis ZT, Boundy-Mills K, Mills DA. A new perspective on microbial landscapes within food production. Current Opinion in Biotechnology 37 (2016):182–189.
- Mayo B, Rachid CTC, Alegría Á, Leite AM, Peixoto RS, Delgado S. Impact of next generation sequencing techniques in food microbiology. Curr. Genom 15 (2014):293-309.
- Ercolini D. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Applied and Environmental Microbiology 79 (2013): 3148-3155.
- Kelleher P, Murphy J, Mahony J, van Sinderen D. Next-generation sequencing as an approach to dairy starter selection. Dairy Sci Technol 95 (2015): 545-568.
- Doyle CJ, Gleeson, D., O’Toole P, Cotter PD. High-throughput metataxonomic characterization of the raw milk microbiota identifies changes reflecting lactation stage and storage conditions. International Journal of Food Microbiology 255 (2017): 1-6.
- Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep 6 (2016): 19233.
- Po?ka J, Rebecchi A, Pisacane,V, Morelli L, Puglisi E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiology 46 (2015): 342-56.
- Murphy J, Bottacini F, Mahony J, Kelleher P, Neve H, et al. Comparative genomics and functional analysis of the 936 group of lactococcal Siphoviridae phages. Sci Rep 6 (2016): 21345.
- Ortea I, O'Connor G, Maquet A. Review on proteomics for food authentication. Journal of Proteomics 147 (2016): 212-225.
- Kergourlay G, Taminiau B, Daube G, Champomier Verges MC. Metagenomic insights into the dynamics of microbial communities in food. Int J Food Microbiol 213 (2015): 31-39.
- Piotrowska-Cyplik A, Myszka K, Czarny J, Ratajczak K, Kowalski R, Biega?ska-Marecik R, Staninska-Pi?ta J, Nowak J, Cyplik P. Characterization of specific spoilage organisms (SSOs) in vacuum-packed ham by culture-plating techniques and MiSeq next-generation sequencing technologies. J Sci Food Agric 97 (2017): 659-668.
- Alkema W, Boekhorst J, et al. Microbial bioinformatics for food safety and production. Briefings in Bioinformatics 17 (2016): 283–292.
- Kumar A and Chordia N. Bioinformatics Approaches in Food Sciences. J Food Microbiol Saf Hyg 2 (2017).
- Cantarel BL, Coutinho PM, Rancurel C, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37 (2009): D233–8.
- Brul S, Schuren F, Montijn R, Keijser BJF, van der Spek H, Oomes SJCM. The impact of functional genomics on microbiological food quality and safety. International Journal of Food Microbiology 112 (2006): 195-199.
- Esposito A, Colantuono C, Ruggieri V and Chiusano ML. Bioinformatics for agriculture in the Next-Generation sequencing era. Chem Biol Technol Agric 3 (2016).
- Reinhardt JA, Baltrus DA, Nishimura MT, Jeck WR, Jones CD, Dangl JL. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res. 19 (2009): 294–305.
- Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, et al., Sequencing and beyond: integrating molecular omics for microbial community profiling. Nature Reviews Microbiology 13 (2015): 360–72.
- Sirangelo TM. Food Microbiology and Multi-Omics Approaches. International Journal of Advances in Science Engineering and Technology 6 (2018).
- Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Bioliology 18 (2017): 83.
