Integrative In-Silico Evaluation of Features on BRCA1 Cis Regulatory Element
Article Information
Apeksha Arun Bhandarkar, Smeeta Shrestha*
School of Basic and Applied Sciences, Dayananda Sagar University, Bangalore, India
*Corresponding author: Smeeta Shrestha, School of Basic and Applied Sciences, Dayananda Sagar University, Bangalore, India
Received: 16 June 2021; Accepted: 28 June 2022; Published: 04 July 2022
Citation: Apeksha Arun Bhandarkar, Smeeta Shrestha. Integrative In-Silico Evaluation of Features on BRCA1 Cis Regulatory Element. Journal of Bioinformatics and Systems Biology 5 (2022): 93-107.
Share at FacebookAbstract
Genomic cis regulatory elements support the gene transcriptional landscape which fine tune spatiotemporal gene expression via interaction with different transcription factors and co modulators during development. These regulatory elements are poorly conserved, highly heterogenous with limited understanding of their role in gene expression. Here we use a well-known human tumor suppressor gene, Breast Cancer Type 1 (BRCA1) and UCSC human genome browser database to report the in-silico putative cis regulatory enhancer element and its features. We report a 2kb double elite enhancer, GH17J043079 located within intron 12 of the BRCA1 gene. The enhancer interacts with NBR1, NBR2, TMEM106A and RPL27 and VAT1 gene promoters. GH17J043079 showed histone activity in human embryonic stem cells, cancerous cells, housed transcription factors specific to liver cells and was enriched with Alu elements, indicative of ability for potential gene rearrangements. Additionally, it contained eQTLs, rs4793197, rs8176190, rs8176192, rs8176193 and rs8176194 with disparity in allele frequency across populations. Our in-silico review on the features present within GH17J043079 element in BRCA1 helps to postulate an intricate transcription regulation. Such candidate based analysis of features within cis regulatory element on a gene can help elucidate intricate genomic architecture, gene regulation and its impact on complex disorders.
Keywords
BRCA1; Sporadic Breast Cancer; Enhancer; SNPs; Transcription Factors; Alu Elements
BRCA1 articles; Sporadic Breast Cancer articles; Enhancer articles; SNPs articles; Transcription Factors articles; Alu Elements articles
BRCA1 articles BRCA1 Research articles BRCA1 review articles BRCA1 PubMed articles BRCA1 PubMed Central articles BRCA1 2023 articles BRCA1 2024 articles BRCA1 Scopus articles BRCA1 impact factor journals BRCA1 Scopus journals BRCA1 PubMed journals BRCA1 medical journals BRCA1 free journals BRCA1 best journals BRCA1 top journals BRCA1 free medical journals BRCA1 famous journals BRCA1 Google Scholar indexed journals Sporadic Breast Cancer articles Sporadic Breast Cancer Research articles Sporadic Breast Cancer review articles Sporadic Breast Cancer PubMed articles Sporadic Breast Cancer PubMed Central articles Sporadic Breast Cancer 2023 articles Sporadic Breast Cancer 2024 articles Sporadic Breast Cancer Scopus articles Sporadic Breast Cancer impact factor journals Sporadic Breast Cancer Scopus journals Sporadic Breast Cancer PubMed journals Sporadic Breast Cancer medical journals Sporadic Breast Cancer free journals Sporadic Breast Cancer best journals Sporadic Breast Cancer top journals Sporadic Breast Cancer free medical journals Sporadic Breast Cancer famous journals Sporadic Breast Cancer Google Scholar indexed journals Enhancer articles Enhancer Research articles Enhancer review articles Enhancer PubMed articles Enhancer PubMed Central articles Enhancer 2023 articles Enhancer 2024 articles Enhancer Scopus articles Enhancer impact factor journals Enhancer Scopus journals Enhancer PubMed journals Enhancer medical journals Enhancer free journals Enhancer best journals Enhancer top journals Enhancer free medical journals Enhancer famous journals Enhancer Google Scholar indexed journals SNPs articles SNPs Research articles SNPs review articles SNPs PubMed articles SNPs PubMed Central articles SNPs 2023 articles SNPs 2024 articles SNPs Scopus articles SNPs impact factor journals SNPs Scopus journals SNPs PubMed journals SNPs medical journals SNPs free journals SNPs best journals SNPs top journals SNPs free medical journals SNPs famous journals SNPs Google Scholar indexed journals Transcription Factors articles Transcription Factors Research articles Transcription Factors review articles Transcription Factors PubMed articles Transcription Factors PubMed Central articles Transcription Factors 2023 articles Transcription Factors 2024 articles Transcription Factors Scopus articles Transcription Factors impact factor journals Transcription Factors Scopus journals Transcription Factors PubMed journals Transcription Factors medical journals Transcription Factors free journals Transcription Factors best journals Transcription Factors top journals Transcription Factors free medical journals Transcription Factors famous journals Transcription Factors Google Scholar indexed journals Alu Elements articles Alu Elements Research articles Alu Elements review articles Alu Elements PubMed articles Alu Elements PubMed Central articles Alu Elements 2023 articles Alu Elements 2024 articles Alu Elements Scopus articles Alu Elements impact factor journals Alu Elements Scopus journals Alu Elements PubMed journals Alu Elements medical journals Alu Elements free journals Alu Elements best journals Alu Elements top journals Alu Elements free medical journals Alu Elements famous journals Alu Elements Google Scholar indexed journals Complex interactions articles Complex interactions Research articles Complex interactions review articles Complex interactions PubMed articles Complex interactions PubMed Central articles Complex interactions 2023 articles Complex interactions 2024 articles Complex interactions Scopus articles Complex interactions impact factor journals Complex interactions Scopus journals Complex interactions PubMed journals Complex interactions medical journals Complex interactions free journals Complex interactions best journals Complex interactions top journals Complex interactions free medical journals Complex interactions famous journals Complex interactions Google Scholar indexed journals DNA articles DNA Research articles DNA review articles DNA PubMed articles DNA PubMed Central articles DNA 2023 articles DNA 2024 articles DNA Scopus articles DNA impact factor journals DNA Scopus journals DNA PubMed journals DNA medical journals DNA free journals DNA best journals DNA top journals DNA free medical journals DNA famous journals DNA Google Scholar indexed journals GTEx articles GTEx Research articles GTEx review articles GTEx PubMed articles GTEx PubMed Central articles GTEx 2023 articles GTEx 2024 articles GTEx Scopus articles GTEx impact factor journals GTEx Scopus journals GTEx PubMed journals GTEx medical journals GTEx free journals GTEx best journals GTEx top journals GTEx free medical journals GTEx famous journals GTEx Google Scholar indexed journals
Article Details
1. Introduction
Complex interactions between transcription factors, chromatin modifications and cofactors at genomic regulatory elements have been known to mediate transcriptional programming. Regulatory elements such as enhancers contain specific DNA elements that are recognized by tissue-specific transcription factors and help program optimal activation of target genes via long range chromatin interactions [1-3]. Such specific interaction events determine spatiotemporal patterns of gene expression that control the events from development of specific cell type, tissues and responses to environmental stimuli, including onset and development of diseases [4]. Importantly, genetic alternations on enhancer sequences have known to contribute significantly to disease progression [5-8]. Studies have reported ~ 80% genetic variants are associated with complex traits residing in non-coding regulatory regions [9] and disease associated variants are enriched in enhancer regulatory elements [3]. The sizes and number of enhancers linked to a gene reflect its disease pathogenicity [10]. The enhancer signature and landscape contribute significantly to gene regulation, expression, and dys-regulation of genes leading to disease conditions specially in multistep malignant cellular transformation [4]. Multiple enhancers can act in a coordinated fashion to regulate spatiotemporal transcription of one or multiple genes [11]. There is limited information on systematic investigation of DNA regulatory elements which are poorly conserved and highly heterogenous in eukaryotic systems [12]. Histone marks and chromatin states support DNA accessibility but there is still limited documentation on the type of features, its interactions within/between enhancer elements which pivot gene regulation. Numerous computational tools and databases like the Roadmap [13], ENCODE [14], FANTOM [15] and Blueprint/IHEC [16] have been integrated within the UCSC genome browser to facilitate accessibility and identify the regulatory elements [14] including enhancers [17], however its accuracy is still low due to sequence heterogeneity [18]. Apart from above databases, there are Single Nucleotide Polymorphisms (SNPs) databases, which help identify variants within DNA regulatory elements. Any kind of sequence alternations within enhancer elements is crucial since it drives the aberrant regulation of oncogenes in cancer [19].
The BRCA1 is a well-studied human tumor suppressor gene located on chromosome 17, band q21.31, spans across Chr17: 43,044,295-43,125,483 [Build GRCh38/hg38] and plays an important role in maintaining genomic stability [20]. BRCA1 generates 34 transcripts and the protein constitutes of the (a) RING domain, present at the N-terminal (between exon 2 to 7); (b) serine cluster domain, present around exon 11-13 central region of the BRCA1 gene and (c) two BRCT domains, at the C- terminal [exon 16-24], which is a phosphoprotein binding domain with specificity for proteins phosphorylated by ATM/ATR kinases. [21] BRCA1 protein has a significant role in both hereditary and sporadic breast cancers [22]. Hereditary breast cancers constitute ~5-10% [23] and are caused due to an inheritance of gene mutation and is recognized by early age at onset and family history [24]. Sporadic breast cancers arise from gene damage acquired from environmental exposures, dietary factors, hormones, aging, and other influences and account for ~90% - 95% of total breast cancer burden [25]. Sporadic breast cancers are attributed to altered expression of BRCA1 protein [22] and studies report, reduced levels of BRCA1 mRNA in majority of breast cancers [26], suggesting its role on the onset and progression of sporadic breast cancer.
These studies indicate dysregulated BRCA1 gene transcription and a requirement to diagnose gene regulatory features/mechanisms within BRCA1 which may contribute in regulation of gene transcription. Although limited, there are experimental evidences where chromatin state and histone marks determine BRCA1 promoter accessibility and the balance of transcriptional co- activators and co-repressors influence gene expression [27]. Secondly, a study identified a 36 bp positive regulatory region (PRR) within BRCA1 which recruited specific factors and on deletion of PRR there was a significant reduction in BRCA1 transcriptional activity [28]. Thirdly, transposable elements on BRCA1 introns have reported rearrangements [29] which are associated with genetic variation and loss of function leading to pathogenesis [30]. Last but the most studied are the BRCA1 single nucleotide polymorphisms that impact transcriptional regulation, and alter promoter activity [31].
Therefore, reports of features on BRCA1 regulatory elements could give better insight to its expression. Although there are extensive data and tools for investigation of gene transcription, regulation and genetic variation, there is limited information on the how features within DNA regulatory elements can influence factors regulating these genes. There is a need to identify such features within regulatory elements in genes across different data sets within a specific biological context to add valuable information about biological processes. In this review we show BRCA1 as a candidate gene to report features residing within its DNA regulatory element to highlight its landscape with the intention to elucidate a need to use this information in an integrative manner to better target BRCA1 linked sporadic breast cancer.
2. Methods
2.1 ENCODE data mapping to BRCA1
The University of California Santa Cruz (UCSC) Human Genome Browser of genome builds GRCh37/hg19 and GRCh38/hg38, were evaluated at chromosome coordinates chr17: 41,196,312- 41,277,500 and chr17: 43,044,295-43,125,483
respectively. Encyclopedia of DNA Elements (ENCODE) data and tracks was utilized to identify the functional elements on BRCA1.The ENCODE tracks including features, GeneHancer, histone marks (H3K4me1, H3K36me3, H3K9ac, H3K27ac),
chromatin state segmentation, transposable elements, repeat regions, transcription factor binding (TFB) and expressed quantitative trait loci (eQTLs) from GTEx v8 was mapped on BRCA1 genomic region. Tracks specific to cell lines was used to integrate and compare chromatin states and transcription factor binding in the enhancer region [Supplementary Figure S1].
2.2 Single nucleotide polymorphism mapping to BRCA1
The NCBI SNP database (dbSNP) was used to search for SNPs located specifically within the DNA regulatory elements on BRCA1 identified by the ENCODE tracks. These SNPs were analyzed using Ensembl Variant Effect Predictor (VEP) [32] to find the Scale-Invariant Feature Transform (SIFT) [33] and Polymorphism Phenotyping (PolyPhen) [34]. SIFT and PolyPhen were used to identify SNPs which had a pathogenic effect on the regulatory region or protein function due to alteration in amino acid sequence. The SNPs present on regulatory region of BRCA1 were analyzed using Linkage
Disequilibrium haplotype (LDhap), matrix (LDMatrix) and trait (LDtrait) Tools. They were used to report the population specific allele frequencies, measure correlation between SNPs and associate SNPs to traits. SNPs with alleles frequencies > 10% and correlation >50% were reported [35].
3. Results
3.1 Double elite enhancer
We report a 2,654 bp double elite regulatory feature, BRCA1/GH17J043079, which spans the between 13/24 exon and 12/23 intron. Approximately 98% of this double elite, medium to weak regulatory element resides within non coding region of the BRCA1 gene as annotated by the GeneHancer regulatory elements and gene interactions database track [36]. Predicted regulatory elements are annotated “double elite” if their GeneHancer gene association scores are obtained from more than one data source indicating a robust prediction which is likely to be functional.
3.2 Elite enhancer and its interactions
The GRCh38/hg38 genome (2013) build reports the elite enhancer regulatory element at chromosome position chr17:43079182-43081835 and interacts with promoter of Neighbor of BRCA1 gene 1 (NBR1) which further interacts with promoter of Transmembrane Protein 106A (TMEM106A) located at chr17:43210408-43214787, situated downstream to BRCA1 [Figure 1]. The TMEM106A gene codes for a transmembrane protein that activates macrophages, upregulates the expression of CD80, CD69 and MHC II on macrophages and induces the release of pro-inflammatory cytokines. The GH17J043079 enhancer in genome build GRCh37/hg19 (2009) reported to interact with the promoter of the Neighbor of BRCA1 gene 2 (NBR2, chr17:41,277,600-41,292,342) which interacts with promoter of the Ribosomal Protein L27 (RPL27, chr17: 41149342-41154588) gene and Vesicle Amine Transport 1 (VAT1, chr17:41176061-41178050) gene, located upstream to BRCA1 [Figure 2]. RPL27 encodes a member of the L27e family of ribosomal proteins required for catalyzing of protein synthesis and VAT1 gene encodes protein responsible for regulating storage and release of neurotransmitters in the nerve terminal and vesicular transport. NBR2 gene is known as a junk gene which encodes for a long non-coding RNA [36]. The enhancer, GH17J043079 on interacting with the NBR2 promoter, GH17J043124 may influence the transcriptional activity of BRCA1 [37] and/or expression of different gene targets (VAT1 and RPL27) of NBR2.
The human gene database, GeneCards [38] annotated the elite enhancer to interact with long noncoding RNA (NONHSAG021878.2 and
NONHSAG021878.2) and the piwi interacting RNAs (piR-60898-014, piR-48218-015), which have the ability to repress the mobilization of transposable elements (TEs) and maintain genomic integrity via transcriptional or post-transcriptional mechanisms [39] [Supplementary Table S1].
Chromatin dynamics on double elite enhancer Chromatin dynamics reported as chromatin states, histone marks and DNase hypersensitivity sites indicate accessibility of DNA for The double elite enhancer on BRCA1 reports different chromatin states depending on type of cell line. It is reported as weakly active (yellow) in the human embryonic stem cells (hESC) and active (green) in chronic myelogenous leukemia cells (K562), human mammary epithelial cells (HMEC) and human lung fibroblast (NHLF) respectively [Supplementary
Figure S2]. The highly dynamic histone marks report potential DNA regulatory features on the genome. Four cell lines reported histone methylation (H3K4me1, H3K36me3) and acetylation (H3K9ac, H3K27ac) marks on BRCA1/GH17J043079 enhancer in genome build GRChr37/hg19. An average ChIP- seq signal of 4.4 units (SD= 3.2) was observed in human embryonic stem cells (H1-hESC) and 5.9 (SD
=4.9) in hepatocytes (HepG2) for H3K4me1 activity. Similarly, ChIP-seq signal of 7.1 (SD 5.2) and 12.11 (SD=5.7) was observed for H3K36m3 in human erythroleukemic cell line (K562) and HeLa cells respectively indicating active transcription. H3K27ac average value of 5.57 (SD=3.06) in H1-hESC suggest active enhancer activity/function, [Supplementary Figure S3] where the average value indicates the intensity of histone activity across the enhancer region. Overall, the histone marks on GH17J043079 suggest active function in H1-hESC and cancerous cells (K562, HeLa).
3.4 Single nucleotide polymorphism on elite enhancer
Prior studies report genetic polymorphisms within coding and non-coding regions contribute to BRCA1 functioning [40, 41]. SNPs also help illustrate the mechanistic importance of a locus to explain their association with risk for associated traits [42]. The genome build GRCh38/hg38 at chr17: 43079182 to 43081835 reported 612 SNPs on the elite enhancer,
28 were unique and associated with breast cancer pathogenicity. These were intronic variants and were clinically reported as benign: likely benign: likely pathogenic at the ratio 17:10:1. Further annotation of these SNPs identified a splice acceptor variant (rs374435098), missense variant (rs183331660, rs374519494 and rs1555583238) and a frameshift variant (rs1555583250). From a total of 28 SNPs, only rs1555583238 showed pathogenic effect with SIFT value of 0 and PolyPhen value of 0.93 [Table 1].
The SNPs, rs4793197, rs8176190, rs8176192, rs8176193 and rs8176194 demonstrated high variation in their allele frequencies across populations. The major and minor allele frequency ratio of rs4793197 (G/A) in the African (AFR), European (EUR) and Asian (SAS) population were 84/16, 64/35, 50/50, demonstrating a gradual change in rs4793197 allele frequency ratio from 8:2 to 5:5 from African to Asian. The major to minor allele frequency of SNPs rs8176190 (C/T) was, 98/1.7, 81/19 and 68/32; rs8176192 (C/G) was 52/48; 99/0.5 and 97/3.2; rs8176193 (C/T) reported 78/22; 64/36 and 50/50 and rs8176194 (A/C) showed 84/16, 64/36 and 50/50 across AFR, EUR and SAS respectively [Figure-3]. There is a minimum of ~18% and maximum of 35% difference in allele frequencies across the 5 intronic variants present on BRCA/GH17J043079 regulatory element. This gradual increase in the minor allele frequency across different populations may suggest potential genetic drift via either natural selection or mutations.
Allele frequencies of SNPs present on elite enhancer were correlated (R2) across African (AFR), European (EUR) and South Asians (SAS) populations with help of LD Matrix. Majority of the SNPs were highly correlated, however few exceptions showed weak correlation in AFR but moderate (60%) to high (~90%) in SAS and EUR. We observed that correlation of between rs8176193 and rs8176194 in AFR (0.64), SAS (0.98) and EUR (1), was 0.64,0.98 and 1 respectively. Similarly, correlation reported as follows between rs4793197: rs8176194 (R2= 0.989, 0.996, 1), rs4793197: rs8176193 (R2= 0.657, 0.991, 1) and rs8176186: rs8176192 (R2 = 0.017, 0.399, 0.967). Higher correlation across SNPs located on BRCA1/GH17J043079 across population indicated similar allele frequency, while low R2 suggested weak correlation indicative to variations across population [Supplementary Figure S4]. SNPs have been associated with risk of numerous complex disorders via genome wide association studies. LDtrait Tool was used to explore the association between SNPs present on enhancer and any known trait. There was no direct association with any trait, however, we observed enhancer SNPs, rs4793197, rs8176194, rs8176193 and rs8176190 showed 74- 97% correlation with 4 other SNPs which were associated with traits like menopause, menarche, blood protein levels, body mass index and eosinophil counts [Supplementary Table S2].
3.5 Elite enhancer on BRCA1 recruit cell line specific transcription factors (TFs)
Enhancers function to fine tune target gene transcript levels by recruiting transcription factors like CRE binding Protein (CREBP) which further mediates the recruitment of Polymerase II at the gene promoter [43] in a cell type specific gene expression. Three transcription factors, zinc finger protein 384 (ZNF384), hepatocyte nuclear factor 4 alpha (HNF4A) and specificity protein 1 (SP1) in liver cells is shown to bind to the elite enhancer element, BRCA1/ GH17J043079.The transcription factor, ZNF384 binding to GH17J043079 enhancer region, showed a strong binding (dark gray; score = 340) activity in liver carcinoma cells (HepG2), HNF4A, showed 2 medium binding active peaks (score = 202, 195) in liver tissue and SP1, showed low intensity binding (light gray; Score = 133) peak in liver samples. Over all potential binding efficiency of different transcription factors to the elite enhancer element from low to high order is SP1, HNF4A and ZNF384, with light grey for SP1 and dark grey for ZNF384. We also searched TF chip seq data in A549, GM12878, H1-hESC, HECK293T, K562, MCF7 cells but did not find any transcription factor binding to the BRCA1/GH17J043079 enhancer element [Figure 4a].
3.6 eQTLs span the elite BRCA1 enhancer element
The expressive quantitative trait loci (eQTLs) are enriched within enhancer elements [3, 44-46] and facilitate in regulation of gene expression by increasing transcription levels and providing spatiotemporal gene expression [10]. A total of 5 cis - eQTLs, rs8176193, rs8176194, rs4793197, rs8176190 and rs8176192, were present within BRCA1/ GH17J043079 [Figure-5] and surprisingly none of them influenced the gene expression of BRCA1. Instead, the eQTLs influenced the expression of 6 genes proximal to BRCA1, across 28 different tissues types indicated by GTEx [47]. The 6 genes whose expression are regulated by the eQTLs are - NBR2, VAT1, LINC00854, PSMC3IP, RND2, TMEM106A in following different tissues, adrenal gland, artery aorta, artery coronary, brain, colon, esophagus, lung, muscle, pituitary, skin, spleen, stomach, thyroid and testis.
Most of the eQTL report negative effect on expression, indicating that presence of the SNP on the enhancer negatively influences the expression of the genes across the different tissues measured in fragments per kilobase of transcript per million mapped reads (FPKM). The rs8176193, reduced RND2 gene expression in testis (FPKM = -12.612) and rs8176190 reduced VAT1 gene expression in esophagus (FPKM = -5.122). However, eQTLs rs8176194 (FPKM = 1.215) and rs4793197 (FPKM =
1.215) positively influenced TMEM106A gene expression. The rs8176193 moderately downregulated the NBR2 gene expression across adrenal gland (FPKM = -0.713), artery aorta (FPKM= -0.644), artery coronary (FPKM = -0.695), brain (FPKM = -0.823), colon (FPKM = -0.858), lung (FPKM = -0.731), prostate (FPKM = -0.942), skin (FPKM = -0.925), spleen (FPKM = -0.939) and stomach (FPKM = -1.042) tissues. Since the NBR2 and BRCA1 genes have a bi-directional promoter, it can be speculated that an alteration in the transcription level of NBR2 can influences the transcription and expression of BRCA1 indirectly [37].
BRCA1 elite enhancer contain Alu elements The BRCA1 elite enhancer contains 3 short interspersed nuclear elements (SINEs) repeats belonging to the Alu family, AluSp (180bp), AluJo (282bp) and AluSx3 (85bp) and one 59 bp long interspersed element 1 (LINE), L2b [Figure-4b]. These Alu elements, AluSp (SW Score = 1431), AluJo (SW Score =1874) illustrate a stronger signal (dark grey) compared to AluSx3 (SW score =402) and L2b (SW score = 212) (light gray). Lighter gray shade of the signal, having lower Smith Waterman (SW Score) alignment score, indicates more number of base mismatch, base deletion and base insertion. BRCA1 is known to be a hotspot for Alu-Alu recombination and contain 137 Alu’s in its 23 introns which make up 40% of its gene sequence [48]. The presence of a SINEs and LINEs within the BRCA1 elite enhancer can rearrange BRCA1 gene hence altering its functioning. This high density of Alu elements within BRCA1 enhancer region and its recombination events, indicates a direct association with the alteration in genome regulation in breast cancer [49].
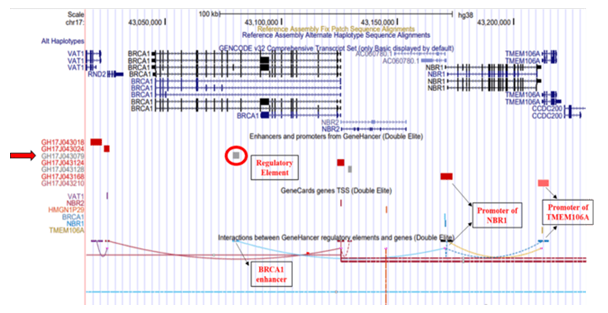
Figure 1: Genome Build GRCh38/hg38 illustrating GH17J043079 regulatory element on BRCA1. The regulatory element (circled in red) is an elite enhancer (GH17J043079) in BRCA1 gene. This enhancer region interacts with the promoter region of NBR1 gene, which further interacts with the promoters of TMEM106A gene.
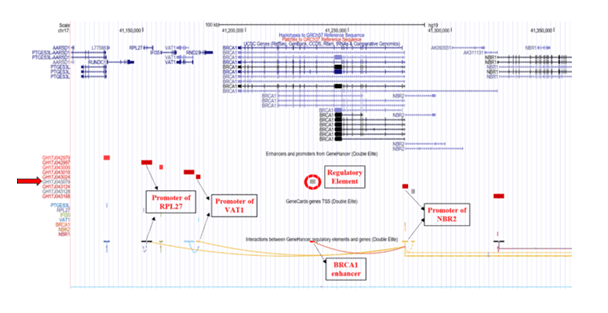
Figure 2: Genome Build GRCh37/hg19 illustrating GH17J043079 regulatory element on BRCA1. The regulatory element (circled in red) is an elite enhancer (GH17J043079) in BRCA1 gene. This enhancer region interacts with the promoter region of NBR2 gene which further interacts with the promoters of VAT1 and RPL27 genes.
Table 1: SNPS present on GH17J043079. The tables report list of filtered SNPs based on clinical relevance, SIFT and PolyPhen values present on the enhancer (GRCh38/hg38) coordinates Ch17: 43079182 to 43081835.
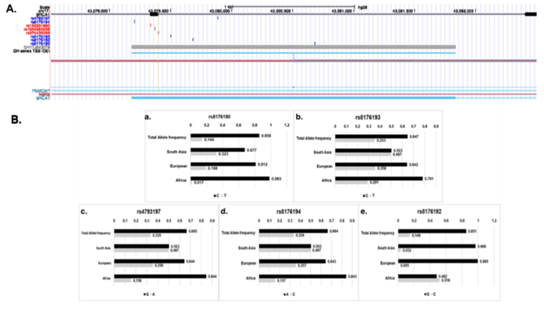
Figure 3: Illustration of SNPs present on GH17J043079 and its allele frequencies across AFR, EUR and SAS. (A) Genomic location of SNPs on GH17J043079 enhancer. Red SNPs indicate protein altering variants and splice site variants and blue indicate intronic variants. (B) Indicates allele frequencies of SNPs, rs8176190, rs8176193, rs4793197, rs8176194 and rs8176192 present on enhancer across African (AFR), Europe (EUR) and South Asian (SAS).

Figure 4: Transcription Factor and Transposable elements on GH17J043079. (A) Shows presence of transcription factors SP1 (light gray – low binding activity), HNF4A (gray- moderate binding activity) and ZNF384 (dark gray – strong binding activity) are present on the enhancer region, which is specific to liver cell and tissue cell line. (B) Short and long interspersed nuclear element (SINE/LINE) transposable elements present on GH17J043079.

Figure 5: Expressed Quantitative trait locus present on GH17J043079. eQTLs, present on enhancer known to regulate 6 genes across 28 tissues.
4. Discussion
We review and report an in-silico analysis of a cis regulatory element, GH17J043079 as a double elite enhancer residing partially in exon 13/24 and intron 12/23 of the BRCA1 gene. It is reported to interact with NBR1 and NBR2 promoters and has further long-distance interactions with TMEM106A gene promoter, RPL27 and VAT1 gene promoters respectively, based on different genome builds. Histone marks, SNPs, transcription factors, transposable elements and eQTLs analysis within the GH17J043079 established it as a regulatory element in the BRCA1 gene. Additionally, presence of histone marks demonstrated high activity of BRCA1/ GH17J043079 in human embryonic stem cells and cancerous cells. We observed that SNPs, rs4793197, rs8176190, rs8176192, rs8176193 and rs8176194 on BRCA1 enhancer element which showed significant variation in allele frequency across ethnicities and rs374435098, a “likely pathogenic” variant was involved in alternate splicing while rs8176193 variant in BRCA1 gene had a risk of breast cancer. Interestingly, the GH17J043079 enhancer also contained eQTLs which are potential regulators of neighbor genes NBR2, VAT1 and RND2 but not the BRCA1 gene in different tissues. Additionally, the enhancer element is rich with Alu elements indicating potential role of gene rearrangements at BRCA1 and contained transcription factor binding sites specific to liver cells and tissues. This in-silico review reports several regulatory features housed within the BRCA1 double elite element, indicating its complex gene transcription regulation.
Sporadic breast cancer (BC) occurs due to somatic mutations arising from defective DNA repair mechanisms and studies indicate reduced BRCA1 levels in them [26]. Numerous studies have investigated the signaling pathways responsible for modulation of BRCA1 expression in sporadic Breast Cancer [37]. Interestingly studies have identified lacunae, suggesting that it is not appropriate to investigate loss of BRCA1 in cells since it does not explain tissue- and sex-specific cancer development in BRCA1 mutation carriers. Instead, it would be interesting to investigate the regulation of tissue and cell type dependent gene transcription of BRCA1 along with genome stability. The latter might illustrate a comprehensive account for BRCA1- dependent tumor suppression in selective tissues [50- 52]. Here we have analyzed the regulatory features present within the double elite enhancer present on BRCA1 gene to review and report its potential in BRCA1 gene regulation and expression. Many studies report that chromatin modification signatures play integral role in chromatin modification, promoter communication and spatio-temporal gene expression. A chromatin conformation study reports mutation in BRCA1, leading to significant loss of H3K27ac-associated super-enhancers in primary mammary epithelial cells, which led to loss of accessibility and binding to GATA rich regions, which is important for regulation of luminal cell fate in mammary gland [53]. Approximately 80% of breast and ovarian cancer predisposition in due to mutations and gene rearrangements in BRCA1 gene [54-56]. Gene rearrangements on BRCA1 due to transposable elements led to frameshift mutation and alternate splicing [57]. Poly (ADP-ribose) polymerase (PARP) inhibitors is one of the successful novel approaches to targeted cancer treatment. Presence of transposable element on BRCA1, led to rearranged, modified BRCA1 which further contributed to reduced Poly (ADP-ribose) polymerase (PARP) inhibitor therapy [29].
The presence of the Alu rich signature at GH17J043079 region has potential for gene rearrangements which could lead to recruitment of different regulatory molecules and transcription factors (TF) leading to aberrant regulation and nonallelic recombination may lead to duplication and deletion of DNA segments [58], which has been associated with disease [59]. Furthermore, there is evidence which report that Alu elements function as regulatory elements which contain histone marks for active chromatin and show tissue specific enrichment for the H3K4me1 mark [60]. Majority of the intronic variants affect splicing which influences gene transcripts and transcription [61] which predisposes individual to multiple human diseases [62-64]. BRCA1 alternative splice variants contribute to genetic diversity and understanding the contribution of these variants help understand role in tumor suppressor [65]. Studies report a BRCA1 intronic variant which causes loss of donor splice site and partial retention of intron 21 in patient transcript demonstrating its pathogenetic role in breast cancer [66]. Although there is no direct evidence supporting the role of the splice acceptor variant rs374435098, but the intron variant is known to be pathogenic. However, studies have identified 2 novel intronic pathogenic variants BRCA1 c.4484 + 3 A > C (rs80358063) and c.5407–10G > A (rs273901767) in BRCA1, which caused complete splicing aberrations and altered the reading frame [67]. Altered splicing by disrupting the splice recognition motif or promoting incorporation of cryptic splice sites lead to pathogenic consequences in the genome [68-70].
The expression quantitative trait loci (eQTLs) indicates variant to be associated with expression. Literature has demonstrated that eQTL, rs9911630 located at 17q21 showed strong effects on the expression levels of BRCA1 and NBR2 and LINC008854 non-coding RNAs [71]. In our in-silico evaluation we observed the following, firstly, the 5 eQTLs on GH17J043079, majorly regulated gene expression of NBR2 located adjacent to BRCA1 across different tissues. Secondly, the allele frequency of these eQTLs varied between 18% to 35% across African, European and South Asian ethnicity, suggestive of underlying natural selection, genetic drift, and gene flow on them. Additionally, reports suggest that genetic variants with extreme allele frequency differences may underlie some human health disparities across populations [72, 73]. Disease susceptible variants are commonly located in cell type specific enhancer and association networks are built on eQTLs associated to traits of clinical or pharmacological relevance [74]. The eQTLs located on BRCA1 enhancer interact with SNPs which are associated directly with traits like menopause, menarche, blood protein levels and body mass index. Furthermore early menarche and late menopause tend to increase the risk of developing breast cancer [75]. The BRCA1 gene activates in association with various transcription factors (TFs) which are associated to the basal transcriptional machinery and transcription-coupled DNA repair processes [76]. The GH17J043079 enhancer demonstrates liver specific transcription by binding to SP1, HNF4A and ZNF384 transcription factors. Such cell line specific activity of enhancer is known to facilitate sub type specific gene expression that are known to program cancer pathogenesis [77].
In this review we cumulatively report and link the features present in the major double elite cis regulatory element on BRCA1 and speculate its putative role in its regulation. Firstly, the GH17J043079 DNA element constitutes a rich landscape of chromatin accessibility signatures, intronic variants, transcription factors, eQTLs and transposable elements. Secondly, it reports various interactions with adjacent genes, indicating that any modification on GH17J043079 landscape can influence multiple genes and tissue expression levels. The major strength of the in-silico study is the elucidation and reporting of the intricate DNA regulatory features present on one of the cis regulatory elements in BRCA1 gene which can help design future functional studies to understand spatio- temporal BRCA1 expression in context of sporadic breast cancer. The limitation of the study is our restriction to only one regulatory feature and absence of experimental data to validate its role in BRCA1 regulation in sporadic breast cancer. The future investigation can involve in-vitro study of mutation of any single feature (Alu, eQTLs, etc) within the GH17J043079 region, diagnose altered transcript and gene expression profile of BRCA1 or its interactions. In summary our review provides a single candidate based in-silico evaluation of the features present on cis regulatory element in BRCA1 which helps to understand the intricate role of BRCA1 in disease conditions and facilitate to better design functional studies to appreciate BRCA1 complex regulation and spatio temporal expression in sporadic breast cancer.
Declaration of Conflicting Interests
The Authors declare that there is no conflict of interest.
References
- Ong CT, Corces VG. Enhancer function: New insights into the regulation of tissue-specific gene Nature Reviews Genetics (2011).
- Spitz F, Furlong EEM. Transcription factors: From enhancer binding to developmental Nature Reviews Genetics (2012).
- Kyle Kai-How Farh, Alexander Marson, Jiang Zhu, et Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature (2015).
- Hans-Martin Herz, Deqing Hu, Ali Shilatifard. Enhancer malfunction in Molecular Cell 53 (2014): 859-866.
- Ning Qing Liu, Menno Ter Huurne, Luan N Nguyen, et The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nat Commun 8 (2017): 14418.
- Lorenzo Pasquali, Kyle J Gaulton, Santiago A Rodríguez-Seguí, et Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet (2014).
- Roland Jäger, Gabriele Migliorini, Marc Henrion, et Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun 6 (2015): 6178
- Ryan N Doan, Byoung-Il Bae, Beatriz Cubelos, et Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167 (2016): 341- 354.e12
- Matthew T Maurano, Richard Humbert, Eric Rynes, et Systematic localization of common disease-associated variation in regulatory DNA. Science 337 (2012): 1190- 1195.
- Wang X, Goldstein DB. Enhancer Domains Predict Gene Pathogenicity and Inform Gene Discovery in Complex Disease. Am J Hum Genet (2020).
- Wittkopp PJ, Kalay Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics (2012).
- Menno P Creyghton, Albert W Cheng, Grant Welstead G, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A (2010).
- Anshul Kundaje, Wouter Meuleman, Jason Ernst, et Integrative analysis of 111 reference human epigenomes. Nature (2015).
- Dunham An integrated encyclopedia of DNA elements in the human genome Nature (2012).
- Marina Lizio, Jayson Harshbarger, Hisashi Shimoji, et Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol (2015).
- José María Fernández, Victor de la Torre, David Richardson, et The BLUEPRINT Data Analysis Portal. Cell Syst (2016).
- Fernández M, Miranda-Saavedra D. Genome- wide enhancer prediction from epigenetic signatures using genetic algorithm-optimized support vector machines. Nucleic Acids Res (2012).
- Hamutal Arbel, Sumanta Basu, William W Fisher, et Exploiting regulatory heterogeneity to systematically identify enhancers with high accuracy. Proc Natl Acad Sci U S A (2019).
- Sur I, Taipale The role of enhancers in cancer. Nature Reviews Cancer (2016).
- Karami F, Mehdipour A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. BioMed Research International (2013).
- Clark SL, Rodriguez A M, Snyder RR, et al. Structure-function of the tumor suppressor Computational and Structural Biotechnology Journal (2012).
- Fraser JA, Reeves JR, Stanton PD, et al. A role for BRCA1 in sporadic breast cancer. Br J Cancer (2003).
- Kleibl Z, Kristensen VN. Women at high risk of breast cancer: Molecular characteristics, clinical presentation and management. Breast (2016).
- van der Groep P, Bouter A, van der Zanden R, et Distinction between hereditary and sporadic breast cancer on the basis of clinicopathological data. J Clin Pathol (2006).
- Romagnolo A, Romagnolo D, Selmin BRCA1 as Target for Breast Cancer Prevention and Therapy. Anticancer Agents Med Chem (2014).
- Thompson ME, Jensen RA, Obermiller PS, et Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet (1995).
- Di L J, Fernandez AG, De Siervi A, et Transcriptional regulation of BRCA1 expression by a metabolic switch. Nat Struct Mol Biol (2010)
- Thakur S, Croce CM. Positive regulation of the BRCA1 J Biol Chem (1999).
- Yifan Wang, Andrea J Bernhardy, Joseph Nacson, et al. BRCA1 intronic Alu elements drive gene rearrangements and PARP inhibitor Nat Commun (2019).
- Ana Peixoto, Manuela Pinheiro, Lígia Massena, et Genomic characterization of two large Alu-mediated rearrangements of the BRCA1 gene. J Hum Genet (2013).
- Leslie J Burke, Jan Sevcik, Gaetana Gambino, et BRCA1 and BRCA2 5′ noncoding region variants identified in breast cancer patients alter promoter activity and protein binding. Hum Mutat (2018).
- William McLaren, Laurent Gil, Sarah E Hunt, et The Ensembl Variant Effect Predictor. Genome Biol (2016).
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein Nucleic Acids Res (2003).
- Ivan A Adzhubei, Steffen Schmidt, Leonid Peshkin, et A method and server for predicting damaging missense mutations. Nature Methods (2010).
- Machiela MJ, Chanock SJ. LDlink: A web- based application for exploring population- specific haplotype structure and linking correlated alleles of possible functional Bioinformatics (2015).
- Simon Fishilevich, Ron Nudel, Noa Rappaport, et al. GeneHancer: genome-wide integration of enhancers and target genes in Database (Oxford) (2017).
- Mueller CR, Roskelley Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Research (2003).
- Gil Stelzer, Naomi Rosen, Inbar Plaschkes, et The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinforma (2016).
- Saito The epigenetic regulation of transposable elements by PIWI-interacting RNAs in Drosophila. Genes and Genetic Systems (2013).
- Omar Alejandro Zayas-Villanueva, Luis Daniel Campos-Acevedo, José de Jesús Lugo-Trampe, et al. Analysis of the pathogenic variants of BRCA1 and BRCA2 using next- generation sequencing in women with familial breast cancer: A case-control study. BMC Cancer (2019).
- Rojano E, Seoane P, Ranea JAG, et Regulatory variants: From detection to predicting impact. Briefings in Bioinformatics (2019).
- Corradin O, Scacheri PC. Enhancer variants: Evaluating functions in common Genome Medicine (2014).
- Catarino R R, Stark A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription Genes and Development (2018).
- Tuuli Lappalainen, Michael Sammeth, Marc R Friedländer, et al. Transcriptome and genome sequencing uncovers functional variation in Nature (2013).
- Kristin G Ardlie, David S Deluca, Ayellet V Segrè, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science (2015).
- Ryan Tewhey, Dylan Kotliar, Daniel S Park, et Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell (2016).
- John Lonsdale, Jeffrey Thomas, Mike Salvatore, et The Genotype-Tissue Expression (GTEx) project. Nature Genetics (2013).
- Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: Transposable elements and disease. Genome Medicine (2009).
- Anwar SL, Wulaningsih W, Lehmann Transposable elements in human cancer: Causes and consequences of deregulationInternational Journal of Molecular Sciences (2017).
- Chang S, Sharan SK. The role of epigenetic transcriptional regulation in BRCA1-mediated tumor Transcription (2013).
- Savage KI, Harkin DP. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS Journal (2015).
- Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene (2006).
- Xiaowen Zhang, Yao Wang, Huai-Chin Chiang, et BRCA1 mutations attenuate super-enhancer function and chromatin looping in haploinsufficient human breast epithelial cells. Breast Cancer Res (2019).
- Teugels E, De Brakeleer S, Goelen G, et De novo Alu element insertions targeted to a sequence common to the BRCA1 and BRCA2 genes. Hum Mutat (2005).
- Preisler-Adams S, Schönbuchner I, Fiebig B, et al. Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German Cancer Genet Cytogenet (2006).
- De Silva S, Tennekoon KH, Karunanayake EH, et Analysis of BRCA1and BRCA2 large genomic rearrangements in Sri Lankan familial breast cancer patients and at risk individuals. BMC Res Notes (2014).
- Montagna M, Santacatterina M, Torri A, et al. Identification of a 3 kb Alu-mediated BRCA1 gene rearrangement in two breast/ovarian cancer Oncogene (1999).
- Häsler J, Strub K. Alu elements as regulators of gene Nucleic Acids Res (2006).
- Reilly MT, Faulkner GJ, Dubnau J, et al. The role of transposable elements in health and diseases of the central nervous J Neurosci (2013).
- Su M, Han D, Boyd-Kirkup J, et al. Evolution of Alu Elements toward Enhancers. Cell Rep (2014).
- Liede A, Karlan BY, Narod SA. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: A review of the literature. Journal of Clinical Oncology (2004).
- Xiaowei Chen, Tuyet-Trinh N Truong, JoEllen Weaver, et al. Intronic alterations in BRCA1 and BRCA2: Effect on mRNA splicing fidelity and expression. Hum Mutat (2006).
- Kwong A, Wong LP, Chan KYK, et Characterization of the pathogenic mechanism of a novel BRCA2 variant in a Chinese family. Fam Cancer (2008).
- Lopez AJ. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation,” Annual Review of Genetics (1998).
- Orban TI, Olah E. Emerging roles of BRCA1 alternative Journal of Clinical Pathology - Molecular Pathology (2003).
- Maria Valeria Esposito, Marcella Nunziato, Flavio Starnone, et A novel pathogenic BRCA1 splicing variant produces partial intron retention in the mature messenger RNA. Int J Mol Sci (2016).
- Wangensteen T, Felde CN, Ahmed D, et al. Diagnostic mRNA splicing assay for variants in BRCA1 and BRCA2 identified two novel pathogenic splicing aberrations. Hered Cancer Clin Pract (2019).
- House AE, Lynch Regulation of alternative splicing: More than just the ABCs. Journal of Biological Chemistry (2008).
- Phillip J Whiley, Lucia Guidugli, Logan C Walker, et Splicing and multifactorial analysis of intronic BRCA1 and BRCA2 sequence variants identifies clinically significant splicing aberrations up to 12 nucleotides from the intron/exon boundary. Hum Mutat (2011).
- Pagani F, Baralle Genomic variants in exons and introns: Identifying the splicing spoilers. Nature Reviews Genetics (2004).
- Yosr Hamdi, Mariem Ben Rekaya, Shan Jingxuan, et A genome wide SNP genotyping study in the Tunisian population: Specific reporting on a subset of common breast cancer risk loci. BMC Cancer (2018).
- Sulovari A, Chen YH, Hudziak JJ, et al. Atlas of human diseases influenced by genetic variants with extreme allele frequency Hum Genet (2017).
- Mao L, Fang Y, Campbell M, et al. Population differentiation in allele frequencies of obesity- associated SNPs. BMC Genomics (2017).
- Branco PR, de Araújo GS, Barrera J, et al. Uncovering association networks through an eQTL analysis involving human miRNAs and Sci Rep (2018).
- Hamajima Menarche, menopause, and breast cancer risk: Individual participant meta- analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol (2012).
- Venkitaraman AR. Functions of BRCA1 and BRCA2 in the biological response to DNA J Cell Sci (2001).
- Wang J, Dai X, Berry LD, Cogan JD, et al. HACER: An atlas of human active enhancers to interpret regulatory variants. Nucleic Acids Res (2019).
Supplementary:
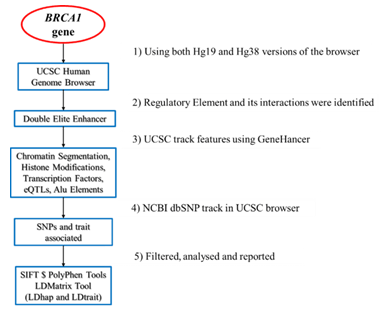
Figure S1: Methodology used to obtain the features of the regulatory element present on BRCA1 gene.
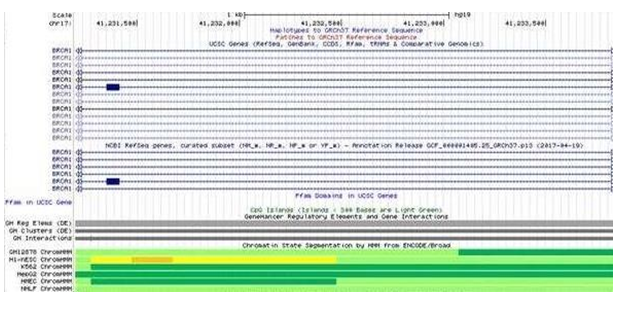
Figure S2: Chromatin Segmentation and Transcriptional Activity of the regulatory element. The yellow region in the hESC cells indicates the regulatory element to be a weak enhancer region. The dark green and the green region in K562, HMEC and NHLF respectively indicates the regulatory element to be a transcriptionally active region.
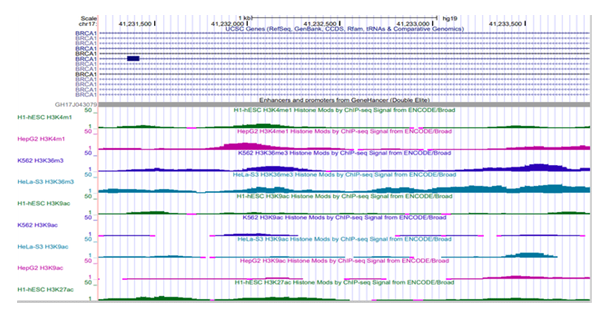
Figure S3: Histone Modification - methylation (H3K4me1, H3K36me3) and acetylation (H3K9ac, H3K27ac) marks on BRCA1/GH17J043079 enhancer region across cell lines.
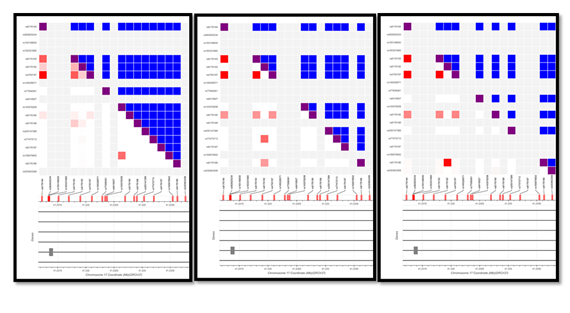
Figure S4: Correlation across the SNPs on BRCA1/GH17J043079 from dbSNP using LD matrix, from left to right in populations of AFR, EUR and SAS. Red squares indicate R2, while the blue dots indicate D’. The bright red dots indicate the correlation > 50%.
|
SL No |
Symbol |
Description |
Function |
|
1 |
BRCA1 |
BRCA1 DNA Repair Associated |
Protein Coding |
|
2 |
RPL27 |
Ribosomal Protein L27 |
Protein Coding |
|
3 |
PSMC3IP |
PSMC3 Interacting Protein |
Protein Coding |
|
4 |
VAT1 |
Vesicle Amine Transport 1 |
Protein Coding |
|
5 |
RND2 |
Rho Family GTPase 2 |
Protein Coding |
|
6 |
NBR2 |
Neighbor of BRCA1 LncRNA2 |
RNA Gene |
|
7 |
PTGES3L-AARSD1 |
PTGES3L-AARSD1 Readthrough |
Protein Coding |
|
8 |
RPL21P4 |
Ribosomal Protein L21 Pseudogene 4 |
Pseudogene |
|
9 |
NONHSAG021878.2 |
LncRNA |
RNA Gene |
|
10 |
piR-48218-015 |
piRNA |
RNA Gene |
|
11 |
NONHSAG021873.2 |
LncRNA |
RNA Gene |
|
12 |
piR-60898-014 |
piRNA |
RNA Gene |
Table S1: Interactions of regulatory element (GH17J043079) with RNA genes. This enhancer region interacts with the RNA genes such as long non-coding RNA and piwi interacting RNAs.
|
Sn No |
Query |
GWAS Trait |
RS Number |
Position (GRCh37) |
R2 |
D' |
|
1 |
rs4793197 |
Body mass index |
rs2670854 |
chr17:41085683 |
0.23 |
0.53 |
|
2 |
rs4793197 |
White blood cell count |
rs28678167 |
chr17:41172481 |
0.38 |
0.85 |
|
3 |
rs4793197 |
Age at menopause |
rs9915489 |
chr17:41173226 |
0.26 |
0.97 |
|
4 |
rs4793197 |
Body mass index |
rs8176166 |
chr17:41240277 |
0.48 |
1 |
|
5 |
rs4793197 |
Eosinophil counts |
rs8176166 |
chr17:41240277 |
0.48 |
1 |
|
6 |
rs4793197 |
Menopause (age at onset) |
rs1799949 |
chr17:41245466 |
0.79 |
0.9 |
|
7 |
rs4793197 |
Menarche (age at onset) |
rs1799949 |
chr17:41245466 |
0.79 |
0.9 |
|
8 |
rs4793197 |
Menopause (age at onset) |
rs8176071 |
chr17:41278005 |
0.87 |
0.99 |
|
9 |
rs4793197 |
Blood protein levels |
rs74252763 |
chr17:41402020 |
0.82 |
0.94 |
|
10 |
rs8176190 |
White blood cell count |
rs28678167 |
chr17:41172481 |
0.14 |
0.81 |
|
11 |
rs8176190 |
Age at menopause |
rs9915489 |
chr17:41173226 |
0.1 |
0.96 |
|
12 |
rs8176190 |
Body mass index |
rs8176166 |
chr17:41240277 |
0.74 |
0.93 |
|
13 |
rs8176190 |
Eosinophil counts |
rs8176166 |
chr17:41240277 |
0.74 |
0.93 |
|
14 |
rs8176190 |
Menopause (age at onset) |
rs1799949 |
chr17:41245466 |
0.39 |
0.98 |
|
15 |
rs8176190 |
Menarche (age at onset) |
rs1799949 |
chr17:41245466 |
0.39 |
0.98 |
|
16 |
rs8176190 |
Menopause (age at onset) |
rs8176071 |
chr17:41278005 |
0.36 |
0.99 |
|
17 |
rs8176190 |
Blood protein levels |
rs74252763 |
chr17:41402020 |
0.37 |
1 |
|
18 |
rs8176190 |
Heel bone mineral density |
rs9904639 |
chr17:41437020 |
0.14 |
0.6 |
|
19 |
rs8176192 |
Coronary artery disease |
rs9912587 |
chr17:41173086 |
0.11 |
0.46 |
|
20 |
rs8176192 |
Age at menopause |
rs9915489 |
chr17:41173226 |
0.16 |
0.99 |
|
21 |
rs8176192 |
Menopause (age at onset) |
rs1799949 |
chr17:41245466 |
0.13 |
1 |
|
22 |
rs8176192 |
Menarche (age at onset) |
rs1799949 |
chr17:41245466 |
0.13 |
1 |
|
23 |
rs8176192 |
Menopause (age at onset) |
rs8176071 |
chr17:41278005 |
0.14 |
1 |
|
24 |
rs8176192 |
Blood protein levels |
rs74252763 |
chr17:41402020 |
0.12 |
0.94 |
|
25 |
rs8176192 |
Heel bone mineral density |
rs9904639 |
chr17:41437020 |
0.12 |
0.46 |
|
26 |
rs8176193 |
Body mass index |
rs2670854 |
chr17:41085683 |
0.21 |
0.54 |
|
27 |
rs8176193 |
White blood cell count |
rs28678167 |
chr17:41172481 |
0.32 |
0.74 |
|
28 |
rs8176193 |
Age at menopause |
rs9915489 |
chr17:41173226 |
0.23 |
0.86 |
|
29 |
rs8176193 |
Body mass index |
rs8176166 |
chr17:41240277 |
0.42 |
1 |
|
30 |
rs8176193 |
Eosinophil counts |
rs8176166 |
chr17:41240277 |
0.42 |
1 |
|
31 |
rs8176193 |
Menopause (age at onset) |
rs1799949 |
chr17:41245466 |
0.9 |
0.99 |
|
32 |
rs8176193 |
Menarche (age at onset) |
rs1799949 |
chr17:41245466 |
0.9 |
0.99 |
|
33 |
rs8176193 |
Menopause (age at onset) |
rs8176071 |
chr17:41278005 |
0.97 |
0.99 |
|
34 |
rs8176193 |
Blood protein levels |
rs74252763 |
chr17:41402020 |
0.89 |
0.96 |
|
35 |
rs8176194 |
Body mass index |
rs2670854 |
chr17:41085683 |
0.23 |
0.54 |
|
36 |
rs8176194 |
White blood cell count |
rs28678167 |
chr17:41172481 |
0.39 |
0.85 |
|
37 |
rs8176194 |
Age at menopause |
rs9915489 |
chr17:41173226 |
0.27 |
0.97 |
|
38 |
rs8176194 |
Body mass index |
rs8176166 |
chr17:41240277 |
0.47 |
1 |
|
39 |
rs8176194 |
Eosinophil counts |
rs8176166 |
chr17:41240277 |
0.47 |
1 |
|
40 |
rs8176194 |
Menopause (age at onset) |
rs1799949 |
chr17:41245466 |
0.79 |
0.9 |
|
41 |
rs8176194 |
Menarche (age at onset) |
rs1799949 |
chr17:41245466 |
0.79 |
0.9 |
|
42 |
rs8176194 |
Menopause (age at onset) |
rs8176071 |
chr17:41278005 |
0.88 |
0.99 |
|
43 |
rs8176194 |
Blood protein levels |
rs74252763 |
chr17:41402020 |
0.82 |
0.94 |
Table S2: The bold text represents SNPs rs4793197, rs8176190, rs8176193 and rs8176194 associated with traits like menopause, menarche and blood protein levels.
