In Vitro Differentiation of Adipose-Derived Mesenchymal Stem cells into Cardiac Tissue-Linked Progenitor Cell Cluster Using Fibrin Matrix Based Niche
Article Information
Subha Sa, Renu Ramesha, K. Jayakumarb, Lissy K Krishnana*
aDivision of Thrombosis Research, Department of Applied Biology, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Sciences and Technology, Trivandrum, 695012, Kerala, India
bDepartment of Cardiovascular and Thoracic Surgery, Hospital Wing, Sree Chitra Tirunal Institute for Medical Sciences and Technology, Trivandrum, 695011, Kerala, India
*Corresponding Author: Lissy K Krishnan, Senior Grade Scientist G, Division of Thrombosis Research, Department of Applied Biology, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Sciences and Technology, Trivandrum, 695012, Kerala, India
Received: 17 October 2020; Accepted: 28 October 2020; Published: 06 November 2020
Citation: Subha S, Renu Ramesh, K. Jayakumar, Lissy K Krishnan. In Vitro Differentiation of Adipose-Derived Mesenchymal Stem cells into Cardiac Tissue-Linked Progenitor Cell Cluster Using Fibrin Matrix Based Niche. Cardiology and Cardiovascular Medicine 4 (2020): 646-678.
Share at FacebookAbstract
Human adipose-derived mesenchymal stem cells (hADMSCs) transplantation has been widely explored for restoring post-myocardial infarct heart function, with limited success. Whereas the multipotent nature of hADMSC could benefit regeneration, differentiation into undesired lineages upon transplantation could detriment the effects. Therefore, controlled in vitro lineage commitment of hADMSC into cardiac cells, before transplantation offers a promising strategy towards better tissue regeneration. The function of cardiac tissue may require a coordinated action of mixed cell types such as cardiomyocyte-like cells (CMLs), endothelial-like cells (ELCs), and cardiac fibroblast-like cells (CFLs). Therefore, as a preliminary step for developing autologous cell-based therapy, this study derived CMLs, CFLs, and ELCs from hADMSCs of cardiovascular disease (CVD) patients. Specifically designed fibrin-based niche aided the lineage commitment of hADMSCs into different cardiac cells. At least two specific markers of each cell type, both at mRNA and protein level, are demonstrated. The co-culturing of hypoxia-induced additional ELC with co-developed CML-CFL-ELC resulted in further upregulation of cardiogenic and angiogenic markers at the transcriptional level. The study confirms the advantage of multipotency of hADMSCs from CVD patients and the designed niche conditions for obtaining viable mixed population cardiac progenitors for potential benefits in individualized MI regeneration approaches.
Keywords
Myocardial infarction; Cardiomyocyte-like cells; Human adipose-derived mesenchymal stem cells; In vitro pre-differentiation; Extracellular matrix; Autologous cell-based therapy
Myocardial infarction articles; Cardiomyocyte-like cells articles; Human adipose-derived mesenchymal stem cells articles; In vitro pre-differentiation articles; Extracellular matrix articles; Autologous cell-based therapy articles
Article Details
Abbreviations
DAPI - 4′,6-diamidino-2-phenylindole; AcLDL - Acetylated low-density lipoprotein; ADMSCs – Adipose derived mesenchymal stem cell/s; ABAM - Antibiotic-antimycotic; BMP-4 - Bone- morphogenetic protein-4; BSA - Bovine serum albumin; CFs – Cardiac fibroblasts; CFLs - Cardiac fibroblast-like cell/s; CSCs - Cardiac stem cells; CMs - Cardiomyocyte/s; CMLs - Cardiomyocyte-like cell/s; CVDs - Cardiovascular disease; cDNA - Complementary deoxy-ribonucleic acid; ELCs - Endothelial-like cell/s; EGF - Epidermal growth factor; ECM - Extracellular matrix; FBS - Fetal bovine serum; GAPDH - Glyceraldehyde -3-phosphate dehydrogenase; GFs - Growth factor/s; hADMSCs - Human adipose derived mesenchymal stem cell/s; HE - Hypothalamus extract; IPs - Induction protocols/1/2/3; IC-SCRT - Institutional Committee for Stem Cell research and Therapy; IEC - Institutional Ethics Committee; IGF-1 - Insulin-like growth factor-1; MSCs - Mesenchymal stem cell/s; MI - Myocardial infarct/infarction; pADMSCs - Patient – specific adipose derived mesenchymal stem cell/s; PGFs - Platelet growth factors; PCR - Polymerase chain reaction; PDGFRα – Platelet derived growth factor receptor alpha; qRT-PCR - Real-time quantitative reverse transcriptase polymerase chain reaction; RNA – Ribonucleic acid; SCT - Sree Chitra Tirunal; SVF - Stromal vascular fraction; TCPS - Tissue culture polystyrene; TGF β – transforming growth factor β; VEGF-165 - Vascular endothelial growth factor- 165.
1. Introduction
Myocardial infarction (MI) is one of the consequences of CVDs causing morbidity and mortality worldwide [1]. Post MI, the resident cardiac stem cells (CSCs) of the heart enter into senescence, and the intrinsic regenerative potential of the heart fails to replace the damaged myocardium [2]. Assistive devices and replacement surgeries are ineffective in myocardial regeneration; therefore, stem cell transplantation seems interesting [3]. Recent therapeutic interventions have focused on the transplantation of stem cells harvested from different tissues [4] for treatment of CVDs by chemical signaling using growth factors (GFs) and cytokines, inducing differentiation [5,6]. Though autologous induced pluripotent stem cell (iPSC) – based differentiation has been achieved by various researchers, translation into clinics is limited due to the high medical cost and prolonged time needed to reprogram and propagate autologous iPSCs. Moreover, it is also reported that iPSCs show greater diversity due to epigenetic memory, genetic background, and novel features obtained during reprogramming [7]. For reducing the medical cost, time, and risk of immune rejection using autologous transplantation, the most convenient cells are adipose-derived mesenchymal stem cells (ADMSCs). Harvesting of adipose tissue causes minimum discomfort and patient-specific ADMSCs, (denoted as pADMSCs) with multipotency may be isolated and expanded with minimal cost, biocompatible reagents, and time. In the context of stem cell transplantation for MI regeneration, lack of integration, and coordination with electrical conductivity of neighboring tissue and arrhythmia have been reported as major challenges [8]. Therefore, a promising strategy to improve the therapeutic efficacy of ADMSCs to regenerate and survive in the infarcted myocardium is pre-differentiation into required cardiac lineage cell types in vitro. However, the disease conditions such as hypertension, hyperglycemia, hyperlipidemia, etc. can affect growth potential and multi-differentiation potency (multipotency) of pADMSCs [9]. Therefore, pre- differentiating pADMSCs into cardiomyocyte-like cells (CMLs), cardiac fibroblast-like cells (CFLs), and endothelial-like cells (ELCs) for developing combination therapy using all cell types seem a challenging problem. Growing differentiated cardiomyocytes (CMs) in the culture itself is hard due to the requirement of a specific niche [10]. For CM differentiation using stem cells, 5-azacytidine was used as an inducer [11], with minimum consistency [12]. Planat – Benard and colleagues have reported the spontaneous differentiation of mice stromal vascular fraction (SVF) cells to cardiac-like phenotype upon culturing them on a semi-solid medium with the influence of cytokines and growth factors (GFs) indicating that the chemical cues triggered the differentiation to cardiac lineage [13]. Subsequently, several research groups have reported various strategies to induce cardiomyogenic differentiation of mesenchymal stem cells (MSCs) using a combination of cytokines or GFs with varying success rates [14,15]. Also, the significance of environmental cues from ECM and the neighboring cells have been shown to play pivotal roles in guiding the survival and differentiation of transplanted stem cells in the injured myocardium [4]. However, no attempt to pre-differentiate pADMSCs to a mixed population of vital cell types present in the myocardial tissue is seen in the literature. Based on this knowledge, designing a cell-specific in vitro niche for generating a mixed population of cardiac lineage cells was the major goal of the study. The major highlight of the study is the exploitation of the multipotency of pADMSC by guided differentiation to derive cells such as CML, CFL, and ELC and to assess the co-survival and phenotype maintenance of each cell type upon co-culture. Fibrin based niche with their ability to act as GF reservoir was proposed for supporting pADMSC growth and differentiation. Modification of various protocols aimed to co-differentiate pADMSC to CMLs and CFLs for improved survival of cardiomyocyte-like phenotype in vitro.
2. Materials and Methods
2.1. Isolation and Characterization of pADMSCs
Human adipose tissue was collected from CVD patients; age group 30-65 (SCT/IEC/453/February 2013; SCT/IC-SCRT/02/February 2013) undergoing coronary artery bypass grafting. The pADMSC was isolated as described [16] and were expanded in culture. Procedures, reagents, culture medium, antibodies used for characterization (Supplementary Table 1), etc. are given in detail in the supplementary file.
2.2 Effect of Fibrin niche on pADMSC differentiation to CMLs
2.2.1. Preparation of Fibrin matrix
Tissue culture polystyrene (TCPS) dishes coated with fibrin matrix as described earlier was used [17]. Briefly, fibrinogen concentrate of clinical-grade fibrin sealant was reconstituted in water. Diluted (5 IU) thrombin was incubated with TCPS for 1-2h, the excess enzyme was aspirated, the surface was air-dried and a thin layer of (12.5 ul per cm2) diluted fibrinogen (2mg ml-1) containing 20µg fibronectin and exogenous gelatin (0.2%; Sigma Chemicals, USA) was placed and allowed to clot, lyophilized (Modulyo 4K, Edwards, UK) in a sterile atmosphere, and stored at 4°C till used.
2.2.2. Induction of pADMSCs to the cardiac lineage
The fibrin-based matrix and the culture medium supplemented with GFs together constituted the biomimetic niche. Initial experiments compared the induction of pADMSC at a seeding density of 5000 cells/cm2 grown on bare TCPS and fibrin matrix coated TCPS. On both bare and coated culture surfaces, the effect of 2 culture media such as DMEM-LG and RPMI 1640 were compared; making 4 different experiments. In all 4 experiments, specific GF inductions were continued for 24 days. pADMSCs cultured using DMEM-LG medium with 10% heat-inactivated fetal bovine serum (FBS; Gibco, USA; Catalog no. 10270-106, South American origin, Batch No.41Q4046K; FBS was heat inactivated at 56°C for 30 min followed by centrifugation at 3000rpm for 15 min and filtered using 0.22µ syringe cartridge before use) and antibiotic-antimycotic (ABAM) on bare TCPS dish was used as the baseline (un-induced pADMSCs) for comparing the relative gene expressions. The RNA was isolated on 8th day, 16th day, and 24th day to quantify the relative gene expressions of cardiac-specific markers as compared to pADMSC grown for each period on bare TCPS.
2.3. Comparison of protocols for co-differentiation into CMLs and CFLs
The co-differentiation to CML and CFL lineages were compared using 3 induction protocols (IPs). In all 3 cases, 1x104 cells/cm2 were seeded on a fibrin-based matrix and cultured for 24h in DMEM/LG medium. The GF induction protocols-designated IP1, IP2, and IP3 were initiated after 24h as described below. Either basic medium RPMI 1640 (Gibco, USA) supplemented with 10% heat-inactivated FBS & ABAM denoted as BM-RPMI1640 or basic medium DMEM/F12 (Gibco, USA) supplemented with 10% heat-inactivated FBS & ABAM denoted as BM-DMEM/F12 was used at different stages of induction/culture. In all 3 cases, cells were analyzed on the 8th day and 16th day of culture for the upregulation of CML and CFL markers using qRT-PCR and immunostaining.
2.3.1. Induction Protocol 1(IP1)
The pADMSC differentiation was initiated by an exchange of medium with BM-RPMI1640 supplemented with 5ng/ml of bone morphogenetic protein-4 (BMP-4; R & D systems, USA). After 48h, BMP-4 was withdrawn and the cells were grown in BM-RPMI1640 supplemented with 5ng/ml Vascular Endothelial growth factor-165 (VEGF-165; Cell Signalling Technology, USA) for 72h. From the 120th hour of initiating induction, culture was continued in BM-RPMI till the 8th/16th day of initiation, at 37°C under 5% CO2 with medium change on alternate days.
2.3.2. Induction Protocol 2 (IP2)
The pADMSC differentiation was initiated by an exchange of medium with BM-RPMI1640 supplemented with a combination of 5ng/ml of Activin A (Invitrogen, USA) and 5ng/ml of BMP-4. After 48h, Activin A and BMP-4 were withdrawn and the cells were supplemented with 5ng/ml VEGF-165 for 72h. From the 120th hour of initiating induction, culture was continued in BM-RPMI-1640 till the 8th/16th day of initiation, at 37°C under 5% CO2 with medium change on alternate days.
2.3.3. Induction Protocol 3 (IP3)
The pADMSC differentiation was initiated by the exchange of medium with BM-DMEM/F12 containing 1.2µg/ml in-house prepared platelet growth factors (PGF) [18], 50ng/ml of Insulin-like growth factor-1 (IGF-1, Cell Signalling Technology, USA) and 50 µg/ml of L-ascorbic acid (Sigma, USA) and after 48h, GFs were withdrawn. After 72h of initiating induction, culture was continued in BM-DMEM/F12 supplemented with 50 µg/ml L-ascorbic acid till the 8th/16th day of initiation, at 37°C under 5% CO2 with medium change on alternate days.
2.4. pADMSC induction to ELC lineage
For pADMSC induction to ELCs in vitro, TCPS was coated as in # 2.2.1 with fibrin, modified by adding in-house prepared hypothalamus extract (HE) [19]. To start with, 1x104 cells/cm2 were seeded on fibrin matrix and were grown in basic medium MCDB-131 (Gibco, USA) supplemented with 10% heat-inactivated FBS (Gibco, USA) and ABAM (Invitrogen, USA) denoted as BM-MCDB-131. After 24h, the medium was exchanged with BM-MCDB-131, supplemented with 50ng/ml VEGF-165, 20ng/ml IGF-1, 10ng/ml recombinant human epidermal growth factor (EGF), (Cell Signalling Technology, USA), heparin sulfate (Sigma), and L-glutamine (Sigma) and was grown for 48h. After 48h, the medium was changed with BM-MCDB-131 and the cells were placed in a hypoxia (5% O2) incubator chamber for 6h (Stem Cell Technologies, USA). The culture was moved to normal culture conditions and cells were analyzed after 24h for gene expression at mRNA level.
2.5. Effect of ELC on CML-CFL upon co-culture
The objective of this part was to identify an additional benefit of paracrine factors from different cell types upon co-culture of hypoxia induced ELCs with CMLs-CFLs. After the withdrawal of hypoxia, ELCs were allowed to grow in culture for 24h, then harvested by trypsinization, and 5000 cells/cm2 were seeded on IP3 induced cells (#2.3.3) on the fifth day of induction. The co-culture was maintained in BM-DMEM/F12-L-ascorbic acid mixed with a 1:1 ratio of BM-MCDB-131 medium supplemented with L-glutamine, L-ascorbic acid, and heparin sulfate till the 8th/16th day of initiating differentiation, at 37°C under 5% CO2 with medium change on alternate days. The cells were analyzed for marker expressions on the 8th day and 16th day of starting CML/CFL/ELC inductions.
2.6. Analysis of differentiation
2.6.1. Real-Time Quantitative reverse transcriptase – PCR
In all cases, RNA was isolated using the TRIZOL reagent (Invitrogen, USA) based on the manufacturer's protocol. RNA quantification was done using Nanodrop 1000 Spectrophotometer (Thermo Scientific, USA). 200ng of total RNA was converted to cDNA using the OrionX cDNA kit (Origin, India) in a thermal cycler (Master cycler; Eppendorf). CML and CFL specific markers were analyzed by qRT-PCR, in a total volume of 15µl containing 20ng cDNA, 100 pM of respective intron spanning forward and reverse primers, and 7.5 µl of OrionX 2X Real-time PCR master mix (Origin, India). Forty cycles of reaction were performed using the Bio-Rad iQ5 Real-time PCR detection system (Bio-Rad, USA) under the following conditions: Enzyme activation, 95°C for 15 minutes; denaturation, 95°C for 30 seconds; annealing, 50°C for 20 seconds; and extension, 72°C for 20 seconds. GAPDH was used as the house-keeping gene. Melt curve analysis was performed for each gene to confirm the specificity of each reaction. PCR products were further analyzed by agarose gel electrophoresis to confirm the amplicon size. The relative gene expression was calculated using un-induced pADMSCs for respective periods and fold change in expression was calculated after normalization with GAPDH expression on each day of analysis using the formula 2-ΔΔCt. The list of primers used for the study is given in Table 1.
|
Gene |
Gene Bank No. |
Primer sequence (5’-3’) |
Amplicon size |
|
GAPDH |
NM_002046.6 |
Forward -GAA ATC CCA TCA CCA TCT TCC AGG Reverse - GAG CCC CAG CCT TCT CCA TG |
120bp |
|
TNNT 2 |
NM_000364 |
Forward - GTT ACA TCC AGA AGA CAG AG Reverse - CTT CAT TCA GGT GGT CAA T |
114bp |
|
MYL 2 |
NM_000432.3 |
Forward - ATT CTC AAC GCA TTC AAA GT Reverse -CAT CTG GTC AAC CTC CTC |
120bp |
|
MYH 6 |
NM_002471.3 |
Forward - GAC CGA GAA TGG AGA GTT Reverse - CAT TTG CTG GGT ATA AGA GAG |
94bp |
|
GJA 1 |
NM_000165 |
Forward - GTC TGA GTG CCT GAA CTT Reverse - GCA CCA CTC TTT TGC TTA A |
97bp |
|
DDR 2 |
NM_001014796 .2 |
Forward - AGT GCC ATC AAG TGT CAA TA Reverse - CGA GTG TTG CTG TCA TCA |
167bp |
|
Fibrillin 1 |
NM_000138.4 |
Forward -TGA TGG CTC CTA CAG ATG TGA ATG C Reverse - GAC ACG GCT GGC AAG GTT CC |
206bp |
|
Collagen 1 |
NM_000088.3 |
Forward - CCA AGG GTA ACA GCG GTG A Reverse - GCT TTC CTT CCT CTC CAG CA |
120bp |
|
CD 31 |
NM_000442.4 |
Forward - CAG TCA TTA CGG TCA CAA T Reverse - CTG AGG ACA CTT GAA CTT C |
118bp |
|
VCAM-1 |
NM_001078.4 |
Forward – CCT CCT TAA TAA TAC CTG CCA TTG Reverse – TCTGTGCTTCTACAAGACTATATGA |
90bp |
|
MCP-1 |
NM_002982.4 |
Forward – CCG AGA GGC TGA GAC TAA C Reverse – ATG AAG GTG GCT GCT ATG A |
120bp |
|
eNOS |
NM_000603 |
Forward – GGC ATC ACC AGG AAG AAG A Reverse – TCG GAG CCA TAC AGG ATT G |
116bp |
|
vWF |
NM_000552.4 |
Forward – CAC CAT TCA GCT AAG AGG AGG Reverse – GCC CTG GCA GTA GTG GAT A |
310bp |
|
pAI |
NM_000602.4 |
Forward – TTG GTG AAG GGT CTG CTG TG Reverse – GGC TCC TTT CCC AAG CAA GT |
485bp |
|
tPA |
NM_033011.3 |
Forward – ATG GGA AGA CAT GAA TGC AC Reverse – GAA AGG GGA AGG AGA CTT GA |
319bp |
|
TGF β I |
NM_000660.6 |
Forward – AGT TGT GCG GCA AGT GGT TGA Reverse – GCC ATG AAT GGT GGC CAG GT |
152bp |
|
PDGFR α |
NM_001347829 .1 |
Forward – AGG TTG AGA GGA GGA CTT Reverse – CCA CTG AGA TGC TAC TGA G |
146bp |
Table 1: Primers used in the study and amplicon size of the products
2.6.2. Immune-specific cell identity
The lineage-specific marker expressions at the translational level for CML, CFL, and ELC markers were analyzed by immunocytochemistry after 16 days of culture. Briefly, the cells were fixed with 3.7% paraformaldehyde (Merck, Germany) for 20 minutes at room temperature, permeated using 0.2% Triton X 100 (Sigma Aldrich, USA) for 5 minutes, and blocked with 3% bovine serum albumin (BSA; Sigma, USA) for 30 minutes. The cells were then incubated with primary antibodies at 4°C overnight, secondary antibodies at 37°C for one hour at room temperature, and stained for nucleus using DAPI (Life Technologies, USA). The differentiation of pADMSCs to ELCs was also analyzed by acetylated Low-density lipoprotein (AcLDL) uptake assay by incubating the induced cells with 10µg/ml AcLDL (Molecular Probes, USA) for 4h in the dark in the incubator at 37°C with 5% CO2. The source of antibodies used for the immunostaining is listed in Table 2. Antibody dilutions in each case are shown in the respective figure legends.
|
Sl.No. |
Name |
Isotype |
Source |
Catalog No. |
|
1 |
Connexin-43 (Cx 43/GJA 1) |
Rabbit IgG |
Thermo Scientific |
PA5-11632 |
|
2 |
Cardiac Troponin T (TNNT 2) |
Mouse IgG |
Abcam |
ab8295 |
|
3 |
Collagen I |
Mouse IgG |
Abcam |
ab6308 |
|
4 |
Fibrillin 1 |
Mouse IgG |
Novus Biologicals |
NB110-8146 |
|
5 |
VCAM-1 |
Rabbit IgG |
Abcam |
ab215380 |
|
6 |
CD31 |
Mouse IgG |
Biolegend |
303110 |
|
7 |
eNOS |
Mouse IgG |
Abcam |
ab76198 |
|
8 |
VE-Cadherin |
Mouse IgG |
Santa Cruz Biotechnology |
sc52751 |
|
9 |
Anti-Mouse AF 488 |
Goat IgG |
Abcam |
ab150113 |
|
10 |
Anti-Mouse TR |
Goat IgG |
Abcam |
ab6787 |
|
11 |
Anti-Rabbit AF 488 |
Goat IgG |
Abcam |
ab150077 |
Table 2: List of antibodies used for the analysis of lineage commitment
2.6.3 Quantification of cardiac Troponin T+ve CMLs
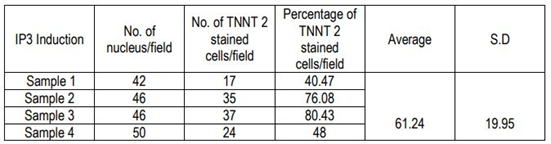
The percentage of cardiac Troponin T positive cells was estimated semi-quantitatively by Image J analysis. The number of nuclei per field was counted from 4 different fields from different donors and the distribution of cardiac Troponin T positive cells per field was counted. The percentage of Troponin T+ve cells was estimated based on the total number of cells in 4 fields and total TNNT 2 cells in the corresponding fields. The average and standard deviation was also calculated.
2.6.4 Analysis of ECM deposition
To determine ECM deposition, cultures were subjected to decellularization, as standardized earlier by Divya et al., (2007) [17] and stained using specific antibodies. Briefly, culture dishes were treated with a solution containing 0.2% NaOH (Merck, Germany), and 1% Triton X – 100 (Sigma Aldrich, USA) in double-distilled water for 30 min. The recovered insoluble matrix was fixed with 3.7% formaldehyde (Merck, Germany) for 20 min and washed. The culture dishes were then incubated with primary antibody against Collagen I (Abcam, UK) for 2h at 25oC and then with Alexa Fluor 488 conjugated secondary antibody (anti-mouse IgG Abcam, UK). Immunostained Collagen I was imaged using a fluorescent microscope (Leica, Germany).
2.7. Statistical Analysis
All experiments were performed at least 3 independent times using different donor cells each time. Statistical significance was calculated by one-way ANOVA (Analysis of Variance) for all quantitative data to determine the significance in variation between control and test. Mean values and standard error were calculated for all parameters.
3. Results
3.1. pADMSC characterization and proof of multipotency
Cells isolated from the patient's adipose tissue were plastic–adherent acquired typical morphology (Supplementary figure S1A), and showed mesenchymal stem cell (MSC) characteristics with surface marker expressions (Supplementary figure S1B) and multipotency (Supplementary figure S1C a-c). Thus, pADMSCs were found suitable for differentiation experiments to derive lineage-committed cardiac cells.
3.2. Influence of Fibrin niche on pADMSC differentiation
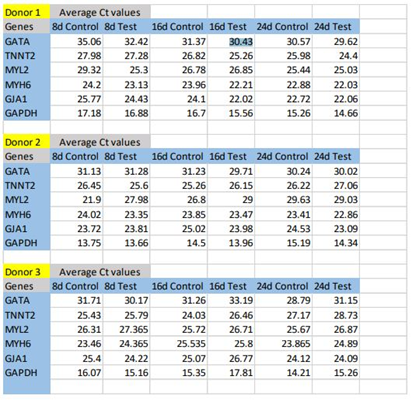
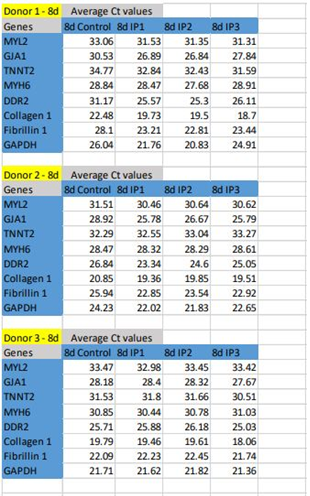
Upon comparison of bare TCPS and fibrin matrix coated TCPS, morphological changes combined with the up-regulation of cardiac-specific genes was observed in the cells cultured on fibrin matrix coated TCPS, indicating that fibrin-based niche may act as a GF reservoir and adhesive protein matrix to derive CMLs from pADMSCs (Supplementary figures S2 to S5). By the 24th day, the pADMSC derived CMLs had reduced marker expressions, indicating that the induced CMLs may need a better niche for phenotype maintenance/differentiation. The average Ct values of cardiac-specific genes analyzed by qRT-PCR represented graphically in Supplementary Figure S5 is provided in the Supplementary Table 2.
3.3. pADMSCs differentiation to CMLs, CFLs & ELCs
3.3.1. Morphological changes
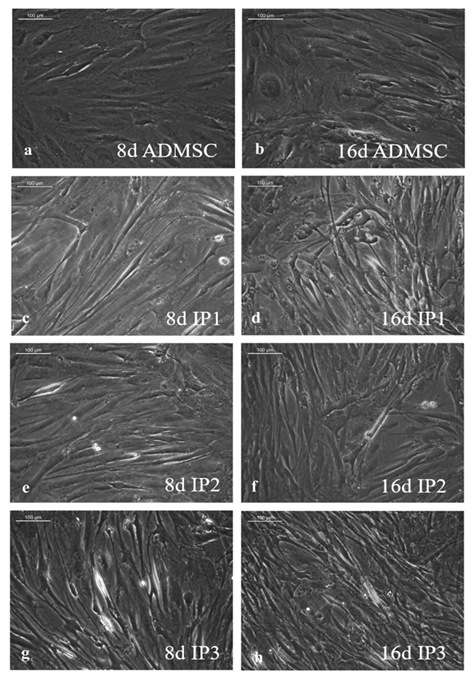
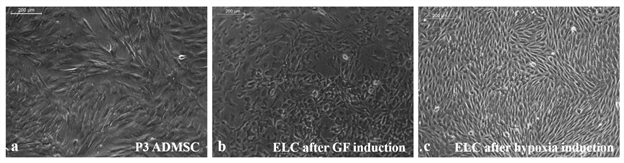
As compared to un-induced pADMSCs, morphological changes were observed in all 3 modes of cardiac inductions. The common changes observed were elongation of cells, the appearance of branching, occasional phase-bright cells, and frequent fibroblastic morphology from the 8th day of differentiation (Figure 1). The pADMSCs induced to ELCs also attained a distinct parallel alignment upon GF induction & uniform cobblestone morphology after exposure to hypoxia (Figure 2).
Figure 1: Representative images showing morphological features of cells under differentiation: 1a, b, Phase contrast image of un-induced pADMSC morphology cultured on bare TCPS on 8d and 16d of culture in vitro, respectively. 1c, d, Phase contrast morphology of pADMSCs induced using IP1 on 8d and 16d of culture in vitro, respectively. 1e, f, Phase contrast morphology of pADMSCs induced using IP2 on 8d and 16d of culture in vitro, respectively. Figure 1g & h, phase contrast morphology of pADMSCs induced using IP3 on 8d and 16d of culture in vitro, respectively. (Magnification – 20X).
3.3.2 mRNA-level indications of differentiation
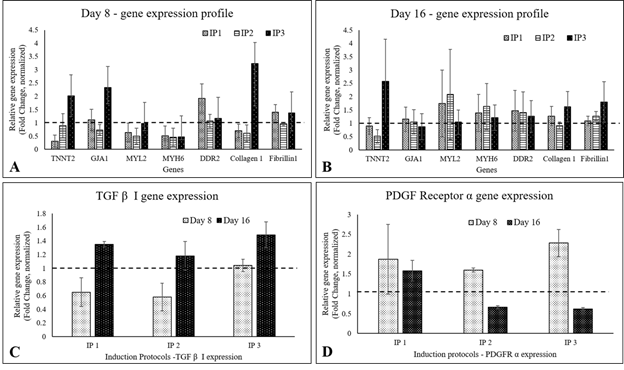
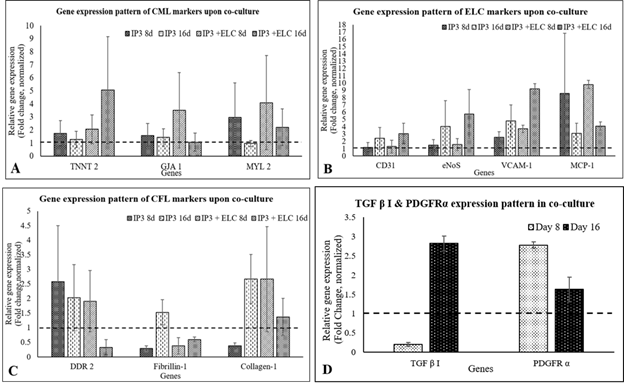
In qRT-PCR, MYL2, TNNT2, and GJA1 were up-regulated in IP3 by 8d of initiating differentiation, while MYH6 gene expression was below 1 fold in all three protocols. By 16th d, the presence of CMLs in IP3 was evident with 1.5- fold up-regulation of TNNT2. The CFL markers DDR2 and Fibrillin-1 were upregulated on 8d and 16d of induction in IP3. Collagen 1, the major ECM marker was found to be 2- fold up-regulated on 8d and 0.5-fold by 16d of IP3 induction, and fibrillin-1 was steady at ~0.5 fold up regulated status (Figure 3). Considering a balance of marker expressions, IP3 seemed to be the most suitable one to derive both CML and CFL simultaneously from pADMSCs. In both IP1 and IP2, TNNT 2 was static on the 8th nor 16th day as compared to un-induced pADMSCs (≤1 fold). The expression of Transforming growth factor β I (TGF β I) and platelet-derived growth factor receptor α (PDGFRα) were also analyzed and in all three IPs, TGF β I expression was low on the 8th day of culture as compared to ADMSC cultured for 8 additional days, whereas its expression was up-regulated by the 16th day (Figure 3C). On the other hand, the expression of PDGFRα was up-regulated by the 8th day of induction while its expression was found to get reduced by the 16th day (Figure 3D). (The average Ct values of lineage specific genes analyzed by qRT-PCR, represented graphically in Figure 3A and 3B is provided in the Supplementary Table 5).
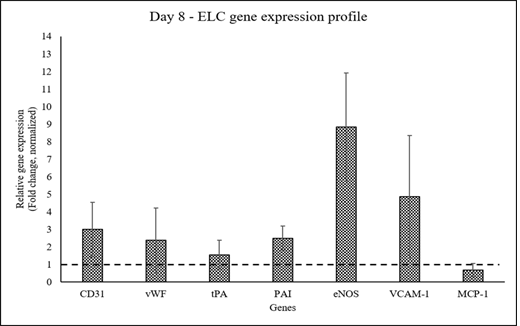
The expression of endothelial cell lineage markers was up-regulated by 8d of pADMSC induction with sequential GF-hypoxia treatment. All ELC-specific genes were up-regulated >1- fold, except MCP-1 on the 8th day of induction indicating lineage commitment of pADMSCs to ELCs (Figure 4).
Figure 3: Graphical representation of the relative gene expression pattern of CML and CFL specific genes by qRT-PCR. The expression pattern of CML and CFL specific genes upon induction of cells using IP1, IP2, and IP3 at (A) 8d and (B) 16d of culture in vitro. The expression pattern of TGF β I, and PDGFRα of cells induced using three induction protocols is represented in Figure 3C and 3D, respectively. Fold change is indicated relative to GAPDH expression on each day of analysis using the 2-∆∆Ct method, upon normalization with normal pADMSCs. The dotted lines represent the basal level expression of genes in un-induced pADMSC cultures on respective periods. Error bars represent standard error (n=3).
Figure 4: Graphical representation of the relative gene expression pattern of pADMSC induced to ELCs. The expression pattern of endothelial lineage-specific markers upon growth factor induction followed by hypoxia treatment. Fold change is indicated relative to GAPDH expression on each day of analysis using the 2-∆∆Ct method, upon normalization with results obtained from the un-induced pADMSCs. The dotted line represents the basal level expression of endothelial-specific genes in uninduced pADMSCs. Error bars represent standard error (n=3).
3.3.3. Identification of functional proteins
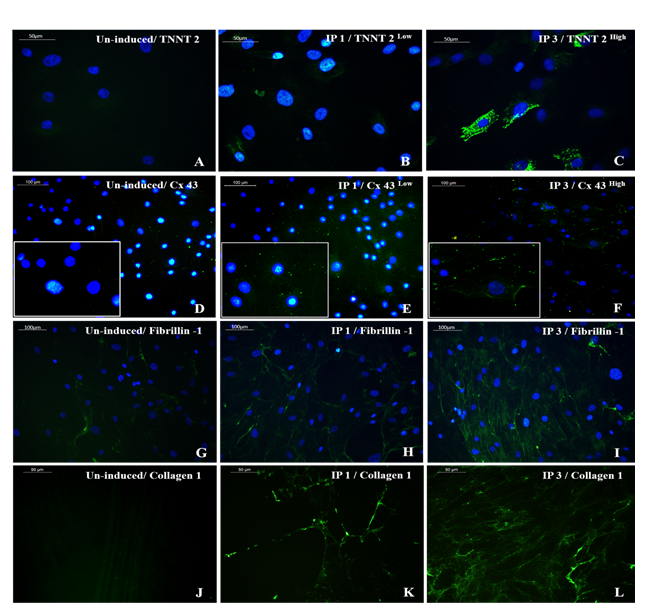
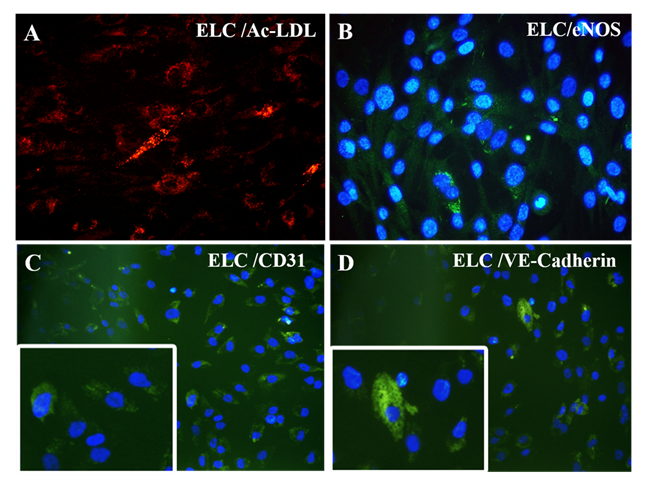
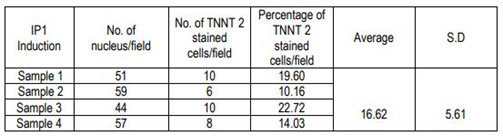
The immune-stained cardiac Troponin T and gap junctional protein Connexin 43 expression were evident in cells induced in the IP3 niche (Figure 5C and 5F, respectively) as compared to weakly stained cells induced in IP1 (Figure 5B, E, respectively), and the observation correlated well with the qRT-PCR data. Upon estimation of cardiac Troponin T expressing cells, around 61.24 ± 19.95% cells in culture were differentiated to CMLs in IP3 (Supplementary Table 4); whereas, in IP1, around 16.62 ± 5.61% only were transformed into CMLs (Supplementary table 3; p = 0.02). The relative distribution of Troponin T+ve CMLs is represented in Supplementary Figure S6. Further, the expression of Fibrillin-1, a cardiac ECM molecule produced by CFLs was expressed in all groups (Figure 5H, I). Cardiac ECM marker, collagen 1 was more prominent in pADMSCs induced using IP3 as compared to IP1 (Figure 5L and 5K, respectively). The endothelial lineage commitment of pADMSCs induced to ELCs was confirmed by AcLDL uptake (Figure 6A) and expression of endothelial cell-specific markers CD31 and VE-Cadherin (Figure 6C and 6D, respectively). eNOS was present in the ELC-specific culture niche with co-localization of blue DAPI of nucleus and green of eNOS making a cyan color in the nucleus and green granular stain in the cytoplasm (Figure 6B). In ELC culture, more than 90% of cells were showing a cyan-colored nucleus; therefore, the majority of cells were differentiated into an endothelial lineage.
Figure 5: Representative images showing immunocytochemical expression of CML and CFL markers. Cardiac Troponin T (TNNT 2) expression (green) was prominent in cells induced using IP3 (5C) while a low expression pattern was observed in cells induced with IP1 (5B) (magnification 40x). Gap junctional protein Connexin 43 (Cx43) was observed in high density (green) along with the membrane structures of cells induced with IP3 (5F) while low expression pattern was observed in cells induced with IP1 (5E) (magnification – 40x). 5H, I represent cardiac fibroblast marker fibrillin-1 in IP1 and IP3 induced cultures, respectively (magnification – 20x); 5K, L represent Collagen 1 staining in decellularized matrix, induced with IP1 and IP3 (green), respectively, on 16d of induction (magnification – 40x). TNNT 2, Cx43, Fibrillin-1, and Collagen 1 staining of un-induced pADMSCs is shown in 5A, 5D, 5G, and 5J respectively. TNNT 2, Cx43, and Collagen 1 were used at 1:100 dilution and Fibrillin-1 was used at a dilution of 1:50.
Figure 6: Characterization of differentiated ELCs: (A) fluorescent micrograph representing AcLDL uptake assay (red); (B) eNOS expression (green in cytoplasm and cyan in nucleus) (magnification – 40x); and (C) CD31 expression (green); and (D) VE-Cadherin expression (green) indicating ELC lineage commitment (magnification – 20x, inset indicating higher magnifications). CD31 and VE-Cadherin were used at a dilution of 1:50 and eNOS at a dilution of 1:100.
3.4. Paracrine effects in co-culture
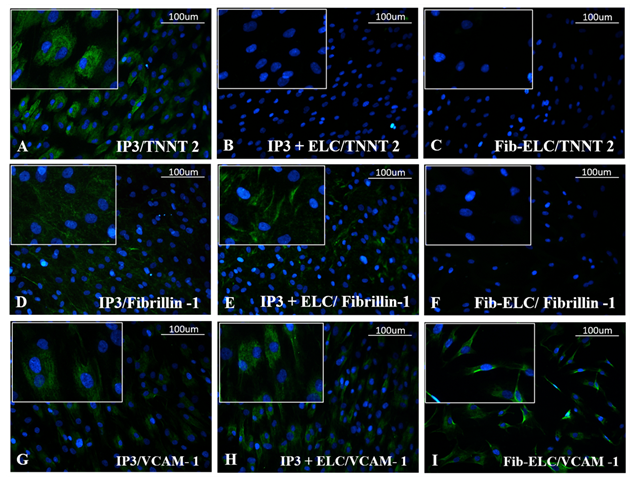
Almost all ELCs adhered to the CML-CFL layer; floating cells were absent in medium (Supplementary figure S7). Upon comparison of gene expression in IP3 with and without ELCs, on 8th day of induction (3rd day of seeding ELCs) & on 16th day, TNNT2, GJA1 and MYL2 were stable, with an evident increase of TNNT2 on the latter period (Figure 7A). On the other hand, DDR2 was down-regulated without much effect on fibrillin 1 and collagen 1 as compared to IP3 alone after similar periods of induction/co-culture (Figure 7C). The ELCs were also stable upon reseeding on IP3, maintenance of CD31, VCAM-1, eNOS, and MCP-1 was observed by the 8th day as compared to ELC before seeding (Figure 4). The expression pattern of endothelial cell (EC) – specific genes were observed further up-regulated by the 16th day upon ELC co-culture with IP3 induced cells (Figure 7B). The gene expression of TGF β I was low on the 8th day while its expression was up-regulated by the 16th day in co-culture. PDGFRα gene expression was up-regulated by the 8th day while its expression pattern was reduced by the 16th day of co-culture (Figure 7D). At the translational level, the expression of cardiac marker TNNT2 and fibroblast marker fibrillin- 1 was observed in the IP3 induced cells on 8d of induction. The development of ELCs in IP3 is evidenced by VCAM-1 stained cells (Figure 8G). Upon co-culture, the ELC layer stained with VCAM-1 was evident, and probable outgrowth of CFL is indicated by Fibrillin-1 (Figure 8H & 8E, respectively). However, CML seemed covered by ELCs and no TNNT2 stained cells were seen (Figure 8B); even though, at the mRNA level, its expression was high in co-culture (Figure 7A).
Figure 7: Graphical representation of relative gene expression pattern of cells upon co-culture. The relative gene expression of (A) CML specific, (B) ELC specific, and (C) CFL specific markers on 8d and 16d of induction normalized to the un-induced pADMSCs is represented graphically. Figure 7D represents the TGF β I and PDGFRα gene expression pattern in co-culture, normalized to the uninduced pADMSCs, respectively. Fold change is indicated relative to GAPDH expression on each day of analysis using the 2-∆∆Ct method, upon normalization with results obtained from the un-induced pADMSCs. Error bars represent standard error (n=3).
Figure 8: Representative immunofluorescent images of CML, CFL, and ELC markers upon co-culture. Cardiac troponin T (TNNT 2) stained images of (A) IP3, (B) IP3 + ELC, and (C) ELC cultures on 8d shown in the top panel. Fibrillin-1 in (D) IP3, (E) IP3 + ELCs and (F) ELC in the middle panel. VCAM-1 immunofluorescence is seen in (G) IP3, (H) IP3 + ELCs, but more prominently in the latter and (I) ELC alone culture on 8d, shown in the bottom panel (magnification -20x). TNNT 2, and VCAM-1 was used at 1:100 dilution and Fibrillin-1 was used at 1:50 dilution (n=3).
4. Discussion
Pathological conditions like MI provoke maladaptive responses in CMs and other non-myocytes resulting in alterations in cardiac function [20]. Owing to inflammation and other structural changes hindering stem cell homing and differentiation, regeneration of damaged myocardium hardly follows the transplantation [2]. The rejuvenation of damaged tissue is further hindered due to the senescent cell pool that gets accumulated in the heart due to disease conditions and aging [21]. Transplantation of stem/progenitor cells is a fascinating area of research towards developing regenerative medicine. In cardiac regenerative approaches, much attention has been given to CMs and endothelial progenitor cell transplantation. Since the CMs co-exist with cardiac fibroblasts (CFs), that represent the major mesenchymal population, and are surrounded by an intricate network of capillaries in cardiac tissue, the interaction of CMs with endothelial cells (ECs) and CFs are essential from the cardiac development stage to the maintenance of intricate cardiac structure and function throughout the life span [22]. They support the myocardium, aiding cell-cell interactions, and 3-dimensional (3-D) architecture of the heart producing autocrine and paracrine action of secreted molecules [23]. Based on such cardiac tissue biology, regeneration of complex cardiac tissue seems to require a heterogeneous population of cells. For coordinating the electrical activity of specialized CM, their survival and functioning require ECM. CFs play a major role in integrating CM beating and electrical conductivity, whereas, angiogenesis and proper oxygenation are essential for the functioning of the tissue.
This study aimed at stem cell priming/pre-differentiation, producing cardiomyocyte-like cells, cardiac fibroblast-like cells, and endothelial-like cells in vitro from single isolation pADMSCs, and conceives potential for future studies towards translation to clinics using less potent patient origin stem cells. Pre-differentiation is an important step in the context of stem cell transplantation because of 2 reasons: (i) MSCs are multipotent and hence may be directed to any lineage depending on signals in the milieu; (ii) the in vivo milieu may not support any differentiation due to inflammatory changes that could deprive adhesive proteins and growth factors. Previous studies have shown that MSCs can be directed into different lineages in vitro via extrinsic growth factor stimulation by the withdrawal of maintenance GFs and the addition of differentiation promoting GFs or chemicals [24]. Behfar et al. have demonstrated that GF combinations including BMP-4, Activin A (members of Transforming Growth Factor-β superfamily), Insulin-like growth factor-1, fibroblast growth factor-2, interleukin-6, and thrombin, induced adult bone marrow-derived mesenchymal stem cells (BMMSCs) into CMs in vitro via endoderm-based pathways by compromising cell proliferation, and switching to differentiation phase [14]. The growth factors BMP-4, Activin A, VEGF-165 for differentiating iPSCs into CMs in vitro [25] and the effect of PGF and IGF-1 in CM differentiation and protection is demonstrated in different studies [22,26,27]. Pretreatment of MSCs with IGF-1 has been shown to improve the healing ability by reducing scar formation and increasing angiogenesis [24]. Sequential treatment of stem cells with growth factor combinations comprising of IGF-1, VEGF-165, and EGF along with heparin sulfate and L-glutamine followed by hypoxia treatment has been shown to induce endothelial lineage commitment. Hypoxia treatment of MSCs is thought to further up-regulate the expression of vascular endothelial growth factor (VEGF) expression in vitro [28].
In the current study, a cell-specific niche comprising fibrin and GFs was utilized effectively; the matrix network functioning as GF reservoir and cell adhesion matrix. Fibrin has been used in bioengineering applications as a scaffold for effective cell attachment, growth, and migration [17] and as a cell delivery vehicle in the treatment of MI [29]. Fibrin comprising various adhesive proteins promotes cell adhesion and proliferation. The fibronectin present in the fibrin matrix acts as a GF reservoir through the heparin-binding repertoire. The major highlight of the study is that, from the same source cells, CML, CFL, and ELC were derived; both CML & CFL were obtained simultaneously in the same niche and culture conditions. The generation of ELC was also indicated in CML/CFL culture in terms of up-regulated EC-specific mRNA expressions; however, to yield significant numbers of ELC expressing specific proteins, a separate protocol was developed. Though a fibrin based niche was developed, specifically to obtain ELC, an additional step of hypoxia exposure made a remarkable improvement in obtaining > 90% ELC phenotype from hADMSCs.
Simultaneous derivation of CFs and CMs from MSCs has not been addressed in earlier studies. This study confirmed that the essential cell category of the myocardium can be obtained in their lineage-committed stage using different GF combinations and culture protocols. The IP3 used a combination of PGF, IGF-1, and L-ascorbic acid on a fibrin-based niche. The composition supported the differentiation and co-survival of pADMSCs into CMLs and CFLs lineages in vitro. The TGF βI was also observed to get up-regulated in the IP3 induction protocol by 16th day, even without the addition of any TGF β superfamily members, indicating that it plays an important role in cardiac differentiation. This observation is in line with previous studies [30-32]. Another observation was the reduced expression of PDGFRα in IP3 by the 16th day of induction. PDGFRα is a well-characterized marker for cardiac progenitor cells and its expression is reported to be minimal in differentiated cardiomyocytes and endothelial cells [33]. The results obtained in our study may thus indicate that the IP3 induction protocol resulted in a progressive differentiation of pADMSCs from a progenitor stage towards a more advanced cardiac lineage-committed phenotype. Though various studies have reported the induction of MSCs using 5- azacytidine, only a minor fraction of ADMSCs was differentiated to cardiomyocytes [34]. Induction of MSCs with either Angiotensin II or 5-azacytidine or a combination of both has been reported to derive cardiomyocyte-like cells upon culturing the MSCs for 3 weeks. However, the stability of derived cardiac progenitors is debated. Exposure to both inducers simultaneously resulted in the death of adherent cells without any favorable additive or synergistic effect on cardiomyocyte differentiation. The estimated yield of cardiomyocytes were; with azacytidine ~24.0%, with Angiotensin II ~25.0%, and with a combination of both ~31% CMLs after 21 days of induced MSC induction [35]. Similarly, a lower frequency of CMLs (20.5 ± 1.9%) was reported to be generated in vitro upon treatment of bone marrow-derived MSCs with TGF-β1 [31]. In this study, the IP3 was found to derive >60% cardiac Troponin T positive CMLs in 16 days period in vitro. This may be probably due to the co-existence of CFLs in the culture. Also, prominent collagen 1 expression in IP3 indicates that by the time fibrin is degraded, collagen-based ECM could form to support cell attachment, growth, and survival. After the grown progenitors were lysed off, uniform distribution of deposited collagen in the culture dish is demonstrated using immunostaining. By this time the adherence of cell to the culture surface was intense which may be due to the collagen deposited by CFL. As a result, single cell suspension, suitable for flow cytometry analysis could not be obtained by standard trypsinization; manual counting was done from multiple fields to estimate % CML cells in the IP3 induced culture. As compared to other reports, the yield of CML in this study is remarkable and the remaining cells could be CFL, which is responsible for collagen deposition and for stabilizing CMLs in the culture. The presence of Connexin 43 in CMLs can promote cell-cell interaction and paracrine signaling for co-survival and functioning. Derivation of dermal fibroblast from hADMSCs has been reported in other studies which included the addition of L-ascorbic acid to promote collagen synthesis and deposition to replace the provisional fibrin-based matrix [36]. However, the major highlight of this study is that in a modified cardiac-specific niche the expression of DDR2, which is a specific cardiac fibroblast molecule that binds to Collagen 1 was obtained. It may be possible that the paracrine factors released from CML influenced the formation of the cardiac phenotype of fibroblasts.
Further, the derivation of ELCs from pADMSCs is confirmed using multiple markers. Even though no specific induction towards ELC was given in IP3, upregulated expressions of ELC markers were detected. Several studies have reported that the co-culture of ECs with CMs promotes survival of the latter population and spatial reorganization [37]. CMs secrete a variety of cardiokines that promote EC activation and proliferation. Similarly, ECs secrete various angiokines that regulate CM proliferation and reorganization following an injury. Hence transplantation of ECs with CMs and CFs could improve the survival of the transplanted cells into an injured heart post-MI [20]. Even though there were detectable levels of EC markers at the mRNA level, at the protein level, not much effect was seen in IP3. Therefore, a separate protocol was developed for generating a stable ELC population from the same donor ADMSCs. The ELC was transferred onto the CML-CFL layer 5 days after primary induction. The cardiomyocytes were not detected at the protein level in co-culture, because the ELCs were not mixed with CML/CFL. The ELCs were transferred and seeded on the IP3 culture surface. It appeared that the CMLs did not sprout out through the ELC layer, whereas, the CFLs outgrew and few fibrillin +ve cells were detected in co-culture, in addition to the EC protein VCAM-1. The co-culture seemed to regulate cell-specific markers of both CML (TNNT2) and CFL (DDR2). The mutual supportive/regulatory role of these 3 cell types is evident in this study. Prospective in vivo experiments assessing the effectiveness of mixed cell phenotypes transplantation by harvesting at early lineage-committed, but proliferative stage may be the next phase of this study. A preliminary study is published [38].
5. Conclusion
In conclusion, this study established that using a single niche and culture conditions pADMSCs can be committed to CML and CFL lineages. Simultaneously, yet another cell-specific niche and culture condition can produce ELC out of pADMSCs. The cell-cell or cell-ECM interactions and the paracrine effects seemed to play a significant role in the maintenance of favorable cell phenotypes in co-culture. The reproducibility of the protocol is proven using different donor cells. The standardized protocol is cost-effective and may be easily adapted for deriving cardiac progenitors from pADMSC in a short time to further develop patient-specific cell-based therapy. The effectiveness of mixed cell transplantation is a relatively less explored concept. The results of this study advocate animal experiments using autologous cells to evaluate the effect of transplantation of mixed progenitor cell populations.
Declarations section
Ethical Approval and Consent to participate
All protocols in this study were approved by the Institutional Ethics Committee (IEC), SCTIMST, (IEC Reference number - SCT/IEC/453/February 2013) and by the Institutional Committee for Stem Cell Research (IC-SCR), SCTIMST (IC-SCR Reference number - SCT/IC-SCRT/02/February 2013) Trivandrum, Kerala, India. Informed consent was obtained from patients before the collection of adipose tissue.
- All authors listed have made substantial contributions as described in the section “Author’s contribution”
- Authors confirm no potential conflicts of interest (financial or non-financial)
Consent for publication
The submission of this manuscript has been approved by all authors. The Director, Head of the Institute has approved the publication of the manuscript, reviewed by the Research and Publication Cell of SCTIMST.
Availability of supporting data
All supporting data has been shown in the current manuscript. The supplementary file is attached separately.
Competing interests
The authors declare no competing financial interests.
Funding
The research fund was received from an internal Institute project (No.6208) and is acknowledged.
Authors’ contributions
Ms. Subha S and Ms. Renu Ramesh equally contributed to this study. Ms. Subha S designed a niche for differentiation of pADMSCs to cardiomyocyte-like cells and cardiac fibroblast-like cells and their molecular and cellular analysis. Ms.Renu Ramesh has standardized a consistent protocol for lineage commitment of pADMSCs to endothelial-like cells and carried out their cellular and molecular characterization. Dr. K. Jayakumar provided patient-specific adipose tissue from CVD patients. Dr. Lissy K Krishnan has contributed to developing the concept and study design, critically revising for important intellectual content, editing, and approving the final version of the manuscript to be published. Authors agree that they are personally accountable for their respective contributions and ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Acknowledgments
The authors are grateful to the Director, SCTIMST, and the Head, Biomedical Technology Wing, SCTIMST for the facilities provided. The authors thank Ms. Priyanka A, Mr. Ranjith S, Mr.Anilkumar V, and Ms. Deepa S for providing the pharmacopeia grade human fibrinogen and thrombin. We thank Dr. Anugya Bhatt for helping with the flow cytometry analysis of ADMSCs. The authors thank SCTIMST for the research fellowship to Subha S and ICMR for the research fellowship to Renu Ramesh. The research fund was received from an internal Institute project (No.6208) and is acknowledged.
Authors’ information
Division of Thrombosis Research, Department of Applied Biology, Biomedical Technology Wing, Sree Chitra Tirunal Institute for Medical Sciences and Technology, Trivandrum 695012, Kerala, India.
Subha S, Renu Ramesh, and Lissy K Krishnan
Department of Cardiovascular and Thoracic Surgery, Hospital Wing, Sree Chitra Tirunal Institute for Medical Sciences and Technology, Trivandrum, 695011, Kerala, India.
Jayakumar K
Corresponding author
Correspondence to Lissy K Krishnan
Supplementary Material
Supplementary Table 1; Supplementary Figure S1; Supplementary Figure S2; Supplementary Figure S3; Supplementary Figure S4; Supplementary Figure S5; Supplementary Table 2; Supplementary Figure S6; Supplementary Table 3; Supplementary Table 4; Supplementary Figure S7; Supplementary Table 5.
References
- Finegold JA, Asaria P, Francis DP. Mortality from ischaemic heart disease by country, region, and age: statistics from World Health Organisation and United Nations. International Journal of Cardiology 168 (2013): 934-945.
- Torella D, Ellison GM, Méndez-Ferrer S, et al. Resident human cardiac stem cells: role in cardiac cellular homeostasis and potential for myocardial regeneration. Nature Clinical Practice Cardiovascular Medicine 3 (2006): S8-S13.
- Ma T, Sun J, Zhao Z, Lei W, Chen Y, Wang X, Yang J, Shen Z. A brief review: adipose-derived stem cells and their therapeutic potential in cardiovascular diseases. Stem Cell Research & Therapy 8 (2017): 1-8.
- Farouz Y, Chen Y, Terzic A, et al. Concise review: growing hearts in the right place: on the design of biomimetic materials for cardiac stem cell differentiation. Stem Cells 33 (2015): 1021-1035.
- Behfar A, Crespo-Diaz R, Terzic A, et al. Cell therapy for cardiac repair—lessons from clinical trials. Nature Reviews Cardiology 11 (2014): 232.
- Li N, Wang C, Jia L, et al. Heart regeneration, stem cells, and cytokines. Regenerative Medicine Research 2 (2014): 1-6.
- Ohnuki M, Takahashi K. Present and future challenges of induced pluripotent stem cells. Philosophical Transactions of the Royal Society B: Biological Sciences 370 (2015): 20140367.
- Goldthwaite Jr CA. Mending a broken heart: Stem cells and cardiac repair. Regenerative Medicine 6 (2006).
- Huang XP, Sun Z, Miyagi Y, et al. Differentiation of allogeneic mesenchymal stem cells induces immunogenicity and limits their long-term benefits for myocardial repair. Circulation 122 (2010): 2419-2429.
- Mitcheson JS, Hancox JC, Levi AJ. Cultured adult cardiac myocytes: future applications, culture methods, morphological and electrophysiological properties. Cardiovascular Research 39 (1998): 280-300.
- Rangappa S, Fen C, Lee EH, et al. Transformation of adult mesenchymal stem cells isolated from the fatty tissue into cardiomyocytes. The Annals of Thoracic Surgery 75 (2003): 775-779.
- Safwani WK, Makpol S, Sathapan S, et al. 5-Azacytidine is insufficient for cardiogenesis in human adipose-derived stem cells. Journal of Negative Results in Biomedicine 11 (2012): 3.
- Planat-Benard V, Menard C, André M, et al. Spontaneous cardiomyocyte differentiation from adipose tissue stroma cells. Circulation Research 94 (2004): 223-229.
- Behfar A, Yamada S, Crespo-Diaz R, et al. Guided cardiopoiesis enhances therapeutic benefit of bone marrow human mesenchymal stem cells in chronic myocardial infarction. Journal of the American College of Cardiology 56 (2010): 721-34.
- Hahn JY, Cho HJ, Kang HJ, et al. Pre-treatment of mesenchymal stem cells with a combination of growth factors enhances gap junction formation, cytoprotective effect on cardiomyocytes, and therapeutic efficacy for myocardial infarction. Journal of the American College of Cardiology 51 (2008): 933-943.
- Zuk PA, Zhu MI, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Engineering 7 (2001): 211-228.
- Sreerekha PR, Divya P, Krishnan LK. Adult stem cell homing and differentiation in vitro on composite fibrin matrix. Cell Proliferation 39 (2006): 301-312.
- Resmi KR, Krishnan LK. Protease action and generation of β-thromboglobulin-like protein followed by platelet activation. Thrombosis Research 107 (2002): 23-29.
- Maciag T, Cerundolo J, Ilsley S, et al. An endothelial cell growth factor from bovine hypothalamus: identification and partial characterization. Proceedings of the National Academy of Sciences 76 (1979): 5674-5678.
- Talman V, Kivelä R. Cardiomyocyte—endothelial cell interactions in cardiac remodeling and regeneration. Frontiers in Cardiovascular Medicine 5 (2018): 101.
- Terzic A, Behfar A. Stem cell therapy for heart failure: ensuring regenerative proficiency. Trends in Cardiovascular Medicine 26 (2016): 395-404.
- Hsieh PC, Davis ME, Gannon J, et al. Controlled delivery of PDGF-BB for myocardial protection using injectable self-assembling peptide nanofibers. The Journal of Clinical Investigation 116 (2006): 237-248.
- Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circulation Research 105 (2009): 1164-1176.
- Youssef A, Aboalola D, Han VK. The roles of insulin-like growth factors in mesenchymal stem cell niche. Stem Cells International (2017).
- Ye L, Zhang S, Greder L, et al. Effective cardiac myocyte differentiation of human induced pluripotent stem cells requires VEGF. PloS One 8 (2013): e53764.
- Dai W, Kloner RA. Cardioprotection of Insulin-Like Growth Factor-1 During Reperfusion Therapy: What Is the Underlying Mechanism or Mechanisms?. Circulation. Cardiovascular Interventions 4 (2011): 311-313.
- Shen H, Wang Y, Zhang Z, et al. Mesenchymal stem cells for cardiac regenerative therapy: optimization of cell differentiation strategy. Stem Cells International (2015).
- Lloyd-Griffith C, Duffy GP, O'Brien FJ. Investigating the effect of hypoxic culture on the endothelial differentiation of human amniotic fluid-derived stem cells. Journal of Anatomy 227 (2015): 767-780.
- Christman KL, Fok HH, Sievers RE, et al. Fibrin glue alone and skeletal myoblasts in a fibrin scaffold preserve cardiac function after myocardial infarction. Tissue Engineering 10 (2004): 403-409.
- Gwak SJ, Bhang SH, Yang HS, et al. In vitro cardiomyogenic differentiation of adipose-derived stromal cells using transforming growth factor-β1. Cell Biochemistry and Function: Cellular Biochemistry and its Modulation by Active Agents or Disease 27 (2009): 148-154.
- Mohanty S, Bose S, Jain KG, et al. TGFβ1 contributes to cardiomyogenic-like differentiation of human bone marrow mesenchymal stem cells. International Journal of Cardiology 163 (2013): 93-99.
- Umbarkar P, Singh AP, Gupte M, et al. Cardiomyocyte SMAD4-dependent TGF-β signaling is essential to maintain adult heart homeostasis. JACC: Basic to Translational Science 4 (2019): 41-53.
- Chong JJ, Reinecke H, Iwata M, et al. Progenitor cells identified by PDGFR-alpha expression in the developing and diseased human heart. Stem Cells and Development 22 (2013): 1932-1943.
- Martin-Rendon E, Sweeney D, Lu F, et al. 5-Azacytidine-treated human mesenchymal stem/progenitor cells derived from umbilical cord, cord blood and bone marrow do not generate cardiomyocytes in vitro at high frequencies. Vox Sanguinis 95 (2008): 137-148.
- Xing Y, Lv A, Wang L, et al. The combination of angiotensin II and 5-azacytidine promotes cardiomyocyte differentiation of rat bone marrow mesenchymal stem cells. Molecular and Cellular Biochemistry 360 (2012): 279-287.
- Sivan U, Jayakumar K, Krishnan LK. Matrix-directed differentiation of human adipose-derived mesenchymal stem cells to dermal-like fibroblasts that produce extracellular matrix. Journal of Tissue Engineering and Regenerative Medicine 10 (2016): E546-E558.
- Narmoneva DA, Vukmirovic R, Davis ME, et al. Endothelial cells promote cardiac myocyte survival and spatial reorganization: implications for cardiac regeneration. Circulation 110 (2004): 962-968.
- Subha S, Sachin J Shenoy, Arya Anil, Sabareeswaran A, Deepthi AN, Lissy K Krishnan. Fibrin Hydrogel Aided Cardiac Progenitor Cell Delivery Enhances Regenerative Tendency in Myocardial Infarct Model. Archives of Clinical and Biomedical Research 4 (2020): 513-541.
Supplementary Information
Isolation and Characterization of pADMSCs
Human adipose tissue was collected from CVD patients of the age group 30-65 (SCT/IEC/453/February 2013; SCT/IC-SCRT/02/February 2013) during coronary artery bypass grafting and isolated pADMSC as previously described (1). Briefly, 5g to 10g of tissue was thoroughly washed with sterile Hank's Balanced Salt Solution (HBSS), minced, and treated with 0.15PZ U/ml of Collagenase NB 4 Standard Grade enzyme (Serva Electrophoresis, Germany) for a period of 45-60 minutes at 37°C with continuous shaking. The digested cell suspension was strained (70uM, BD Falcon, USA), centrifuged, washed 2 times, and the cell pellet was re-suspended in low glucose Dulbecco's modified Eagle's medium (DMEM- LG; Gibco, USA) supplemented with 10% Fetal Bovine Serum (FBS; Gibco, USA; Catalog no. 10270-106, South American origin, Batch No.41Q4046K; FBS was heat-inactivated at 56°C for 30 min followed by centrifugation at 3000rpm for 15 min and filtered using 0.22µ syringe cartridge before use) and Antibiotic- antimycotic solution (ABAM; Invitrogen, USA). The cells were seeded in a 25cm2 tissue-culture polystyrene flask (TCPS; Nunc, Denmark) and incubated at 37°C under 5% CO2. The medium was changed at 3-day intervals. Upon reaching ~ 80% confluence, cells were passaged by standard trypsinization protocol using 0.25% Trypsin- EDTA (Invitrogen, USA) for cell expansion.
pADMSCs from passage 2-4 were used for characterization using surface markers as listed in the supplementary table1 and confirmed by tri-lineage differentiation and specific staining.
Supplementary Table 1: List of antibodies used for pADMSC characterization
|
Sl.No. |
Name |
Isotype |
Source |
Catalog No. |
|
1 |
CD 105 PE |
Mouse IgG |
Santa Cruz |
sc-18838 |
|
2 |
CD 90 PE |
Mouse IgG |
BD Pharmingen |
555596 |
|
3 |
CD 73 PE |
Mouse IgG |
Biolegend |
344003 |
|
4 |
CD 14 FITC |
Mouse IgG |
Millipore |
CBL453F |
|
5 |
CD 45 FITC |
Mouse IgG |
Beckman Coulter |
A07782 |
Adipogenic, osteogenic, and chondrogenic differentiation was attempted using StemPro medium (Life Technologies, USA), as per the standard procedure. For adipogenic induction 1×104 cells/cm2 , for osteogenic induction 5×103 cells/cm2, respectively were seeded and cultured in the respective medium for 21 days with the medium change in 2-3 days. For chondrogenic differentiation cell droplets were seeded and grown for 14 days in the respective medium. Differentiation was confirmed by staining with Oil Red O, Alizarin Red S, and Toluidine blue as per the standard protocols.
Supplementary Results
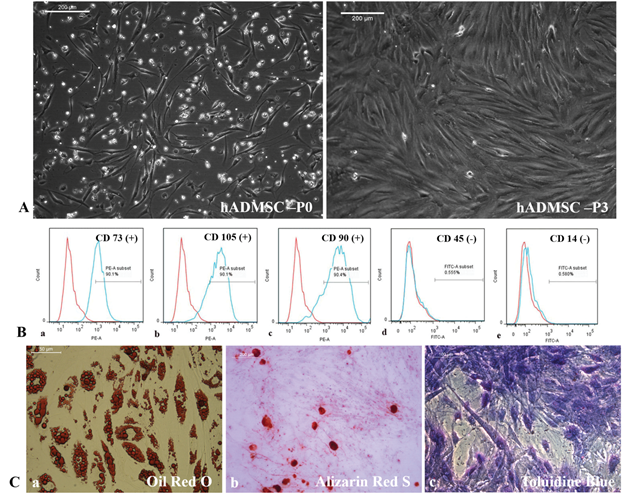
Cells isolated from the patient's adipose tissue were plastic – adherent acquired spindle-shaped morphology from the second day of plating, and became confluent in ~ 8 days. On trypsinization and passaging, every 4 days, the cells became monolayer (Supplementary figure S1A). Flow cytometry analysis of passage 3 (P3) pADMSCs showed 90% positivity for cell surface markers CD105, CD73, and CD90 with < 2% expressing the hematopoietic stem cell markers CD45 and CD14 (Supplementary figure S1B). The multipotent nature pADMSCs were confirmed by adipogenic, osteogenic, and chondrogenic lineage differentiation. Oil Red O staining (Supplementary figure S1C. a), Alizarin Red S Staining (Supplementary figure S1C. b), and Toluidine Blue staining (Supplementary figure S1C. c) confirmed the adipogenic, osteogenic, and chondrogenic lineage commitment, respectively.
Supplementary Figure S1: Morphology and characterization of pADMSCs. S1A, Morphology of pADMSC at passage 0, and passage 3 in TCPS. S1B, Flow cytometry histogram shift of P3 pADMSC characterized using cell surface markers, a) CD73, b) CD105, c) CD90, d) CD45 and e) CD14 as compared to unstained control indicating 90% positivity for CD 73, CD105 and CD90 and < 2% positivity for CD45 and CD14. S1C, Multipotency of pADMSCs indicated by adipogenic differentiation (Oil Red O staining - 40X magnification), osteogenic differentiation (Alizarin Red S staining - 10X magnification), and chondrogenic differentiation (Toluidine blue staining - 10X magnification).
Effect of fibrin coated niche in inducing pADMSCs to cardiac lineage cells
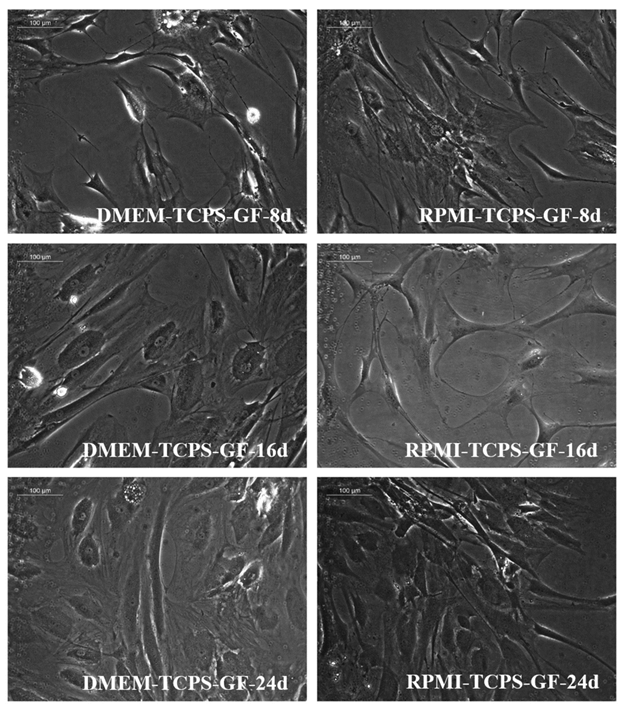
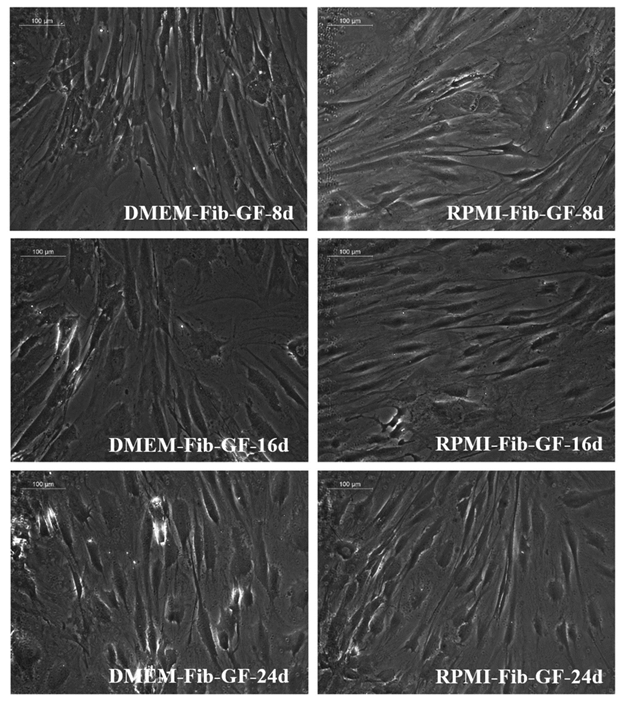
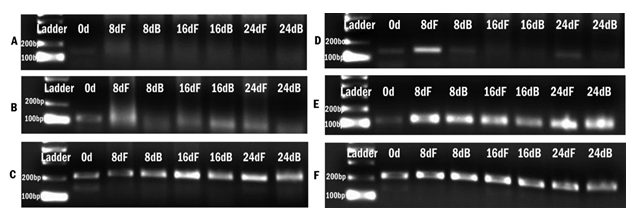
The standardization phase of the differentiation experiment aimed to identify if the fibrin matrix influences pADMSC differentiation into cardiac lineage in 24 days. Morphological changes included elongation of cells, the appearance of cell fusion, and the formation of tube-like structures on fibrin matrix coated culture dishes (Supplementary figure S2 and S3). The effect of fibrin coated niche on pADMSC differentiation to cardiac lineage was further confirmed by reverse transcriptase PCR (RT-PCR) of cardiac-specific markers GATA-4 and Troponin T. Cardiac transcription factor GATA-4 was upregulated in the cells that were induced on the fibrin-based niche as compared to the cells cultured on bare TCPS indicating that fibrin-based niche favored pADMSC differentiation to cardiac lineage in vitro. Also, the expression of GATA-4 and Troponin T was not observed in pADMSCs cultured on DMEM-TCPS and DMEM-Fibrin (Supplementary figure S4). CML marker expression was seen prominent till 16 days in pADMSCs cultured with RPMI-GFs on a fibrin niche, with a decline thereafter (Supplementary figure S5).
Supplementary Figure S2: Phase-contrast images of pADMSCs cultured on bare TCPS. Distinct morphological features are seen depending on the culture medium and period of culture. The left panel indicates cells cultured in DMEM-LG- GFs and the right panel indicates cells cultured in RPMI- GFs. 8d, 16d, and 24d indicate cells cultured for 8 days, 16 days, and 24 days in vitro respectively (magnification – 20x).
Supplementary Figure S3: Phase-contrast images of pADMSCs cultured on fibrin matrix coated TCPS. Distinct morphological features are seen depending on the culture medium and period of culture. The left panel indicates cells cultured in DMEM-LG-GFs and the right panel indicates cells cultured in RPMI- GFs. 8d, 16d, and 24d indicate cells cultured for 8 days, 16 days, and 24 days in vitro respectively (magnification – 20x).
Supplementary Figure S4: Agarose gel images of RT-PCR products of markers expressed upon induction to cardiac cells. A) and D) expression of GATA-4 in cells cultured in DMEM-LG medium and RPMI 1640 medium with GFs respectively. B) and E) expression of TNNT-2 by cells cultured in DMEM-LG medium and RPMI 1640 medium with GFs respectively and C) and F) expression of GAPDH by cells cultured in DMEM-LG medium and RPMI 1640 medium with GFs respectively. 0d indicates day 0; 8dF, 16dF and 24dF indicate gene expression of cells cultured till day 8, day 16, and day 24 on fibrin matrix and 8dB, 16dB, and 24dB indicates cells cultured till day 8, day 16, and day 24 on bare TCPS.
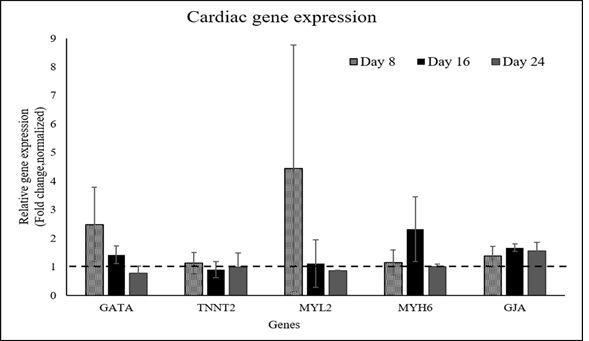
Supplementary Figure S5: qRT-PCR analysis data of CML marker expression in induced pADMSCs grown on a fibrin niche. The expression pattern of CML specific genes at 8d, 16d, and 24d of culture in vitro is graphically represented. Gene expression of each period was normalized with un-induced pADMSCs at respective periods of culture. The dotted line represents the basal level expression of cardiac-specific genes in un-induced pADMSCs.
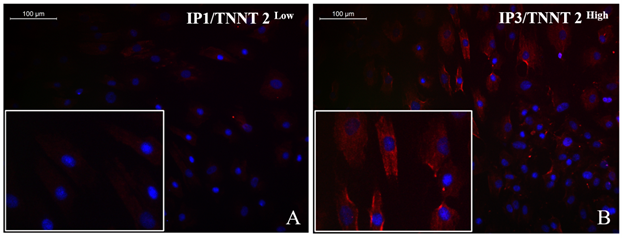
Supplementary Figure S6: Representative fluorescent micrograph images of cardiac Troponin T distribution in induced cells. IP3 induced cells with a more frequent distribution of Cardiac Troponin T (TNNT2) expression (red) (B) as compared to IP1 induced cells (A) indicating more efficient cardiac lineage induction using IP3 induction strategy (magnification – 20x).
References
- Zuk PA, Zhu MI, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Engineering 7 (2001): 211-228.