Identifying the Mutational Profile of ABCB5 Upregulation in Colorectal Cancer: A Comprehensive Database Analysis
Article Information
Rachel E Sexton1, Keerthana Danasekaran1, Husain Y Khan1, Hafiz Uddin1, Mohammad Najeeb Al-Hallak1, Yosef Landesman2, Trinayan Kashyap2, Yiwei Li1, Amro Aboukameel1, Asfar S Azmi1*
1Department of Oncology, Wayne State University, Detroit MI, USA
2Karyopharm Therapeutics Inc., Newton MA, USA
*Corresponding Author: Asfar S Azmi, Department of Oncology, Wayne State University School of Medicine, 4100 John R, HWCRC 732, Detroit MI 48201, USA
Received: 16 July 2021; Accepted: 02 August 2021; Published: 11 August 2021
Citation: Rachel E Sexton, Keerthana Danasekaran, Husain Y Khan, Hafiz Uddin, Mohammad Najeeb Al-Hallak, Yosef Landesman, Trinayan Kashyap, Yiwei Li, Amro Aboukameel, Asfar S Azmi. Identifying the Mutational Profile of ABCB5 Upregulation in Colorectal Cancer: A Comprehensive Database Analysis. Journal of Cancer Science and Clinical Therapeutics 5 (2021): 363-381.
Share at FacebookAbstract
Colorectal cancer (CRC) is a leading cause of cancer related deaths worldwide and the third leading cause of cancer related deaths in the United States with a 5-year survival rate of 63%. Depending on the stage of disease, there are differing rates of overall survival: Stage I (90%) vs. Stage IV (14%). There is a clear discrepancy between early and late stage disease due to a variety of factors including acquired genetic and tumor mutations and lack of initial symptoms leading to late stage detection. It is clear that further understanding into the mechanisms of CRC need to be explored in depth to improve overall survival rates. ATP binding cassette subfamily B member 5 (ABCB5) has been found to be pathogenic in a variety of diseases including melanoma and colorectal cancer but there has been few studies looking at the cellular consequences of ABCB5 upregulation in CRC cells. Using publicly accessible databases, we explored ABCB5 positivity in CRC cells and found novel interactions with 64 differentially expressed genes involved in cellular processes like modulation of inflammation, (XCL1, TNFSF4), enhanced cell signaling via interaction with serine protease inhibitors (SERPINB4, WFDC6) and downregulation of anti-apoptotic proteins (SPIN2A). Although ABCB5 upregulation is not currently targetable with pre-clinical compounds, our analysis revealed the genetic effects induced by ABCB5 upregulation are targetable, including evidence of sensitivity to the XPO1 inhibitor XPOVIO (selinexor). It is clear further pre-clinical investigation is needed to validate these findings.
Keywords
Colorectal cancer; Database analysis; ABCB5; Targetable genes
Colorectal cancer articles; Database analysis articles; ABCB5 articles; Targetable genes articles
Colorectal cancer articles Colorectal cancer Research articles Colorectal cancer review articles Colorectal cancer PubMed articles Colorectal cancer PubMed Central articles Colorectal cancer 2023 articles Colorectal cancer 2024 articles Colorectal cancer Scopus articles Colorectal cancer impact factor journals Colorectal cancer Scopus journals Colorectal cancer PubMed journals Colorectal cancer medical journals Colorectal cancer free journals Colorectal cancer best journals Colorectal cancer top journals Colorectal cancer free medical journals Colorectal cancer famous journals Colorectal cancer Google Scholar indexed journals Database analysis articles Database analysis Research articles Database analysis review articles Database analysis PubMed articles Database analysis PubMed Central articles Database analysis 2023 articles Database analysis 2024 articles Database analysis Scopus articles Database analysis impact factor journals Database analysis Scopus journals Database analysis PubMed journals Database analysis medical journals Database analysis free journals Database analysis best journals Database analysis top journals Database analysis free medical journals Database analysis famous journals Database analysis Google Scholar indexed journals ABCB5 articles ABCB5 Research articles ABCB5 review articles ABCB5 PubMed articles ABCB5 PubMed Central articles ABCB5 2023 articles ABCB5 2024 articles ABCB5 Scopus articles ABCB5 impact factor journals ABCB5 Scopus journals ABCB5 PubMed journals ABCB5 medical journals ABCB5 free journals ABCB5 best journals ABCB5 top journals ABCB5 free medical journals ABCB5 famous journals ABCB5 Google Scholar indexed journals Targetable genes articles Targetable genes Research articles Targetable genes review articles Targetable genes PubMed articles Targetable genes PubMed Central articles Targetable genes 2023 articles Targetable genes 2024 articles Targetable genes Scopus articles Targetable genes impact factor journals Targetable genes Scopus journals Targetable genes PubMed journals Targetable genes medical journals Targetable genes free journals Targetable genes best journals Targetable genes top journals Targetable genes free medical journals Targetable genes famous journals Targetable genes Google Scholar indexed journals tumor mutations articles tumor mutations Research articles tumor mutations review articles tumor mutations PubMed articles tumor mutations PubMed Central articles tumor mutations 2023 articles tumor mutations 2024 articles tumor mutations Scopus articles tumor mutations impact factor journals tumor mutations Scopus journals tumor mutations PubMed journals tumor mutations medical journals tumor mutations free journals tumor mutations best journals tumor mutations top journals tumor mutations free medical journals tumor mutations famous journals tumor mutations Google Scholar indexed journals melanoma articles melanoma Research articles melanoma review articles melanoma PubMed articles melanoma PubMed Central articles melanoma 2023 articles melanoma 2024 articles melanoma Scopus articles melanoma impact factor journals melanoma Scopus journals melanoma PubMed journals melanoma medical journals melanoma free journals melanoma best journals melanoma top journals melanoma free medical journals melanoma famous journals melanoma Google Scholar indexed journals SERPINB4 articles SERPINB4 Research articles SERPINB4 review articles SERPINB4 PubMed articles SERPINB4 PubMed Central articles SERPINB4 2023 articles SERPINB4 2024 articles SERPINB4 Scopus articles SERPINB4 impact factor journals SERPINB4 Scopus journals SERPINB4 PubMed journals SERPINB4 medical journals SERPINB4 free journals SERPINB4 best journals SERPINB4 top journals SERPINB4 free medical journals SERPINB4 famous journals SERPINB4 Google Scholar indexed journals familial adenomatous polyposis articles familial adenomatous polyposis Research articles familial adenomatous polyposis review articles familial adenomatous polyposis PubMed articles familial adenomatous polyposis PubMed Central articles familial adenomatous polyposis 2023 articles familial adenomatous polyposis 2024 articles familial adenomatous polyposis Scopus articles familial adenomatous polyposis impact factor journals familial adenomatous polyposis Scopus journals familial adenomatous polyposis PubMed journals familial adenomatous polyposis medical journals familial adenomatous polyposis free journals familial adenomatous polyposis best journals familial adenomatous polyposis top journals familial adenomatous polyposis free medical journals familial adenomatous polyposis famous journals familial adenomatous polyposis Google Scholar indexed journals peritoneum articles peritoneum Research articles peritoneum review articles peritoneum PubMed articles peritoneum PubMed Central articles peritoneum 2023 articles peritoneum 2024 articles peritoneum Scopus articles peritoneum impact factor journals peritoneum Scopus journals peritoneum PubMed journals peritoneum medical journals peritoneum free journals peritoneum best journals peritoneum top journals peritoneum free medical journals peritoneum famous journals peritoneum Google Scholar indexed journals chemotherapeutic articles chemotherapeutic Research articles chemotherapeutic review articles chemotherapeutic PubMed articles chemotherapeutic PubMed Central articles chemotherapeutic 2023 articles chemotherapeutic 2024 articles chemotherapeutic Scopus articles chemotherapeutic impact factor journals chemotherapeutic Scopus journals chemotherapeutic PubMed journals chemotherapeutic medical journals chemotherapeutic free journals chemotherapeutic best journals chemotherapeutic top journals chemotherapeutic free medical journals chemotherapeutic famous journals chemotherapeutic Google Scholar indexed journals
Article Details
1. Introduction
Within the United States, colorectal cancer (CRC) is the third leading cause of cancer related deaths [1]. In 2020, ~150,000 new cases of CRC were reported which represents 8.2% of all new US cancer cases [2]. Although the 5-year survival rate for localized (Stage I-II) CRC is 90.2%, the 5-year survival rate for metastatic CRC (Stage III-IV) is a mere 14.3% [2]. This survival discrepancy highlights the need for advancements to late stage CRC research. It is well known that early stage CRC has a much better prognosis due to advanced screening methods including commercial detection kits for the at risk population as well as yearly colonoscopies for those over the age of 45. These screening methods have been proven to decrease the likelihood of late stage CRC detection and thus significantly reduce the possibility of further genetic mutations or acquired tumor resistance. Risk factors for developing CRC include, but are not limited to, genetic mutations (such as APC mutation), family history of disease (Lynch syndrome or familial adenomatous polyposis (FAP)), or environmental factors (poor diet, smoking, alcohol abuse). Unfortunately many do not take advantage of these screening methods or have inadequate knowledge of their familial history leading to CRC development commonly caught in its later stages [3-4]. Late stage detection is also attributed to an innocuous group of symptoms including weight loss, abdominal pain and bloody stools, all of which can be attributed to other gastrointestinal conditions. As a result, nearly a quarter of CRCs are diagnosed at advanced stages that present with metastases of the liver, lung and peritoneum [5]. The existing forms of therapy include chemotherapy, radiation therapy and targeted drugs [6]. The most notable chemotherapeutic regimen is the FOLFOX combination, which consists of leucovorin calcium (folinic acid), fluorouracil, and oxaliplatin. Other combinations can be used including CAPOX/XELOX (capecitabine, oxaliplatin), FOLFIRI (Folinic Acid, Fluorouracil, Irinotecan hydrochloride), and FOLFIRI + monoclonal antibodies (bevacizumab or cetuximab) in combination with radiation or alone [7]. Targeted treatments are also being investigated including CAR T cells, synthesized compounds (IMM-101 CPT11), tyrosine-kinase inhibitors (Afatinib, VEGF inhibitors) and small molecule inhibitors (selinexor) (clinicaltrials.gov). Despite therapeutic advancements ~ 90% of metastatic colon cancer, (stage IV CRC) patients acquire resistance to chemotherapy (i.e., 5-fluorouracil, oxaliplatin or Irinotecan) and/or radiation therapy [8].
ATP binding cassette subfamily B member 5 (ABCB5) upregulation was found to be prominent in 5-FU resistant colorectal cancer [9]. ABCB5 spans the plasma membrane and is a P-glycoprotein and ABC transporter [9]. Since this discovery, there has been some work into identifying the functional consequences of ABCB5 upregulation. The phenotypic consequences of ABCB5 upregulation include alterations in critical cancer pathways including growth signaling (such as stimulation of the AXL receptor tyrosine kinase) and drug resistance through ABC transporter upregulation, enhanced glutathione production and have been linked to genetically susceptible melanoma development [10-13]. Although ABCB5 upregulation is known to be prominent in various cancers, there has been little research into the in depth mutational profile and cellular consequences caused by ABCB5 upregulation in CRC. Utilizing publicly available datasets, we aim to explore the in-depth ways in which ABCB5 upregulation is pathogenic to CRC beyond its known functions as well as propose alternative and indirectly targetable strategies within CRC that are induced by cellular ABCB5 upregulation.
2. Materials and Methods
2.1 Cell lines, culture conditions and reagents
HTC-116 and HC-29 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) in 5% CO2 atmosphere at 37 °C (referred to as complete medium). All cells were used up to 25 passages and cell lines were tested and authenticated. Cells were also routinely screened for mycoplasma using PCR analysis.
2.2 CBioPortal database analysis
The cBioPortal for Cancer Genomics was used to obtain survival curves and clinical outcome data for WFDC6 with data relevant to bowel/colon cancer. P-values were calculated with this program. Accessed between 1 April 2021
through 18 May 2021 [14-15].
2.3 Cell cycle arrest
HTC-116 and HC-29 cells were plated at a density of 5 x 105 cells/mL in RPMI without FBS. The following day fresh complete media along with 400 nM selinexor (HTC-116) or 500 nM selinexor (HC-29) were added. Cells were incubated for 24 h then collected and fixed with 70% cold ethanol. Propidium iodide (stock 1 mg/mL) and RNAse I was then added to the cells incubated and submitted to the flow cytometry core at Karmanos Cancer Institute for processing using the BSL-II flow cytometer.
2.4 Growth inhibition by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assay (MTT)
HTC-116 or HC-29 cells were seeded at 1 × 104 cells per well in 96-well micro-titer plate in complete media. The following day the cells were treated with increasing concentrations of selinexor (0-10 uM) taken from 10 mM stock dissolved in DMSO. If dilutions were needed, they were made 1:1000 in PBS. After 72 h of incubation, the MTT assay was performed by adding 20 µL of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma, St. Louis, MO, USA) solution (0.5 mg/mL in PBS) to each well and incubated for 2 h. After the supernatant was aspirated, cells were dissolved in 100 µL of 100% 2-propanol. The plates were rocked for 30 min in a gyratory shaker and absorbance was measured at 570 nm. Results are representative of n=3.
2.5 NCBI GEO database analysis
The NCBI GEO Datasets GSE71670 dataset was utilized for this study comparing ABCB5- positive and ABCB5-negative expression. A box and whisker plot was created through R statistical analysis tool to visualize the data, which appears to be normalized and cross comparable between cells. A moderated T-Statistic was generated through Limma. The moderated t-statistic computed during this test followed the theoretical predicted distribution. All significantly altered genes within GSE71670 were uploaded into the TBtools Toolbox for Biologists software. A volcano plot was produc-ed by this software. The DEGs were selected according to the following criteria: P value < 0.05 and fold change > 1. 248 significant DEGs were found, of the annotated genes 35 were upregulated and 29 were downregulated. The upregulated and downregulated DEGs were entered into DAVID and analyzed through the Functional Annotation Tool. Gene Ontology (GO) analysis for all 64 differentially expressed genes was performed. GO Terms with P value < 0.05 were considered statistically significant [16].
2.6 RNA extraction/RT-qPCR
HC-29 cells were plated at a concentration of 5 x 105 cells/well in 6-well plates. Cells were treated with 500 nM selinexor and incubated for 24 hours. Cells were collected and total RNA was isolated using TRIZOL reagent (Thermo Fisher Scientific). 1-ug samples of cDNA from control vs selinexor treated HC-29 cells were assessed for TROAP using RT-qPCR.
2.7 STRING-dB
STRING-dB was used to identify protein-protein interactions for the following genes: TNFSF8, XAF1, HISTH4D, WFDC6 [17].
2.8 Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
The KEGG Pathway Analysis Tool was used to identify pathways for the differentially expressed genes.
2.9 Drug-Gene interaction database
The Drug-Gene Interaction Database (DGIdb) was used to identify either druggable targets or potential druggable pathways in the differentially expressed genes [18].
2.10 Human protein atlas
The Human Protein Atlas was used to verify the significance of several DEGs in colorectal cancer including SPIN2A, TNFSF8, CCL4, TRAF1, XAF1, ABCB5 and HIST1H4D (http://www.proteinatlas.org) [19-23].
2.11 Statistics
The GEO Database, STRING-dB, Human Protein Atlas, and cBIOPortal provided statistics.
3. Results
3.1 ABCB5 expression upregulates critical pathways in CRC
To begin our investigation into the underlying global molecular aberrations caused by ABCB5 upregulation, we first verified the validity of the GSE71670 dataset in relation to ABCB5-status. We hypothesized that there should be a fold-change difference in ABCB5 expression between the four distinct colon cancer cells lines provided: HT-29 (epithelial-like primary), HTC-116 (epithelial-like, stage unknown), SW620 (metastatic) and COLO741 (metastatic) (Figure 1A). As predicted, we found ABCB5 to be significantly upregulated by a fold change of 43.46 (p-value 0.0000819) confirming that this dataset has a mutual and significantly distinct population of CRCs with highly upregulated ABCB5 expression. CRC patients with low-levels of ABCB5 were found to have significantly better prognostic outcomes (Figure 1B) supporting various literature studied suggesting ABCB5 acts as a cancer-promoting mutation and/or oncogene in cancer. Comparison of the four cell models between ABCB5 positive and negative status revealed 35 upregulated genes, which are involved in immune stimulation, ubiquitination and cell cycle division (Table 1, Figure 1C).
Of the 35-upregulated genes, we were interested in Kinesin Family Member 14 (KIF14), Kinesin Family Member C1 (KIFC1), NDC80 Kinetochore Complex Component (NDC80), and Ubiquitin conjugation enzyme E2 C (UBE2C) as they have been extensively studied in CRC individually, but there has been no connection made between these proteins and ABCB5 expression (Table 1). KIF14 is involved in cell division, mitotic metaphase plate congression and SCF dependent proteasomal ubiquitin-dependent processes. CRC cell proliferation is accelerated by enhancement of the cell cycle through KIF14 activation [24-25]. KIFC1, a kinase receptor, is involved in microtubule-based movement, mitotic sister chromatid segregation, and cell division. In CRC KIFC1 plays an important role in pathogenesis and is involved in cancer stemness [26]. NDC80 is involved in cell division, mitotic sister chromatid segregation, sister chromatid cohesion, mitotic nuclear division, and chromosome segregation. NDC80 was found to be overexpressed in CRC tissue and promotes cancer progression by down regulating tumor suppressor proteins (TSPs) such as FOXO1 [27-28]. Lastly, UBE2C is involved in cell division, regulation of ubiquitin-protein ligase activity, mitotic cell cycle regulation, and proteasome-mediated ubiquitin-dependent protein catabolism. UBE2C is overexpressed in CRC tissue and promotes cell proliferation, invasion, and inhibits apoptosis, which results in the development and metastasis of colorectal cancer [29-30]. Analysis into the differences in CRC disease characteristics between patients harboring mutations in KIFC1, NDC80, KIF14 and UBE2C revealed an increased number of female patients presenting (59.93% vs. 48.37%) as well as a higher abundance of MSI-high status tumors (21.48% vs. 2.13%) (Figure 1D) but alterations in these genes did not correlate with overall survival in CRC patients.
3.2 ABCB5 expression upregulates critical and understudied cellular pathways in CRC
We have identified four novel and upregulated genes within the ABCB5 positive cells (TNFSF8, WFDC6, HIST1H4D and XAF1) that are both underrepresented within the literature and are novel to CRC research to the best of our knowledge (Table 1). Tumor Necrosis Factor Ligand 8 (TNFSF8) is involved with cell-cell signaling, immune responses and positive regulation of RNA Polymerase II transcription [31]. There is a clear and direct link to inflammation and CRC promotion [32]. TNFSF8 activation has been implicated within neovascularization and inflammation on a much smaller scale through a brief mention within a paper suggesting TNFSF15 synergy with IL-12/IL-18 promotes colon cancer [33]. CRC patients were found to have no significant differences in overall survival in relation to TNFSF8 expression but did have statistical significant overall survival differences to proteins which directly interact with TNFSF8 including TRAF1 and CCL4 (Figure 2A). WFDC6, a protease inhibitor, was also found to be upregulated with ABCB5 positivity. This protein is found predominant within semen and the biological processes associated include positive regulation of serine-type endopeptidase activity, regulation of endopeptidase activity, and protein hetero oligomerization. It is interesting that a protein found predominately in semen would be significantly upregulated in four ABCB5-positive CRC models. Regardless, serine proteases play critical roles in tumor invasion and metastasis and further investigation of this protein is needed as mutations in WFDC6 and its directly interacting proteins shows significantly better survival in CRC patients (Figure 2B) [34]. XIAP associated factor 1 (XAF1) was overexpressed in ABCB5 positive cells. XAF1 interacts with ISG15 to promote colon cancer tumor progression and metastasis [35]. XAF1 also interacts with XIAP, a potent member of the inhibitors of apoptosis protein (IAP) family, to inhibit apoptosis [35]. Upregulation of XAF1 caused by ABCB5 upregulation suggests a novel mechanism of CRC survival through suppression of apoptosis, which we postulate, may further contribute to chemotherapeutic resistance. We found CRC patients with low expression of XAF1 has statistically significant higher survival rates in CRC patients conducive with the rationale that XAF1 overexpression is a cancer-inducing protein (Figure 2C). Finally, HIST1H4D is a histone H4 protein involved with nucleosome structure and epigenetic modulation. There has been little research involving HIST1H4D and CRC but its expression has been found to be a prognostic signature in breast cancer [36]. Low expression of HIST1H4D significantly improves overall survival rates in CRC patients indicative of a pathogenic event in CRC (Figure 2D).
3.3 Low ABCB5 expression downregulates anti-cancer survival pathways in CRC
ABCB5 upregulation significantly downregulated 29 genes which are mainly involved in G-protein coupled receptor pathways (GPCRs) such as GPR151 and OR4A15. A handful of small-noncoding RNAs were also found to be downregulated (Table 2). G-protein coupled receptor 151 (GPR151) senses acidic cellular conditions and cellular injury or pain and has been linked to stage II CRC metastatic relapse [37]. Current studies show low expression of GPCR signaling pathways are associated with lymph node metastasis in CRC patients and lower survival rates of CRC patients [38]. If a pre-cancerous or primary malignant colon cancer cell were to acquire upregulation of ABCB5, it can be hypothesized that cellular injury or inflammation caused by this event may have the capability to evade immune detection in part by downregulation of GPR151, but further investigation is needed to confirm. Olfactory Receptor Family 4 Subfamily A Member 15 (OR4A15) is a GPCR that activates olfactory receptors linked to smell. Decreased olfactory receptor activity was found to promote cell proliferation and inhibition of apoptosis, two vital cancer related processes while furthermore olfactory signaling was found to be a new avenue for molecular targets in CRC [39]. Analysis CRC mutations in OR4A15 and other olfactory family members found within the analysis (OR6C3, OR5AK2) significantly altered the pathology of CRC to a hypermuated phenotype (44.19% vs. 3.08%) and shifted the tumor status to a more MSI-high type (32.32% vs. 2.75%) (Figure 3A). SPIN2A was also found to be downregulated in ABCB5 positive cells. Spindlin 2A (SPIN2A) is a tumor suppressor associated with cell cycle regulation and its expression correlates with overall survival in CRC (Figure 3B) [40].
Aside from genetic alterations in ABCB5 upregulated cells, various small-noncoding RNAs were also identified including LIMD1-AS1, SNORD117, SNORD63 and FAM167A-AS1 (Table 2). Out of those, LIMD1-ASI was the only characterized small-noncoding RNA and downregulated within non-small cell lung cancer [41]. It functions naturally as a tumor suppressor via activation of apoptotic processes [41]. Suppression of LIMD1-ASI by ABCB5 upregulation suggests that it functions as a tumor suppressor also within CRC but further work is needed to validate this. Small nucleolar RNAs (SNORDs) found to be downregulated in this study have not been extensively characterized or validated in CRC. What is known is SNORD63 gene location was found to be intronic within heat-shock protein 70 (HSP70), a useful marker for predicting CRC, and has been proposed as a serum diagnostic biomarker for clear cell renal carcinoma [42-43]. SNORD117 gene location was found to be intronic within the spliceosome RNA helicase BAT1, a DEAD box family member that is involved with pre-mRNA splicing and found near immune related genes such as NF-kappa B and TNFa [44].
3.4 Identification of ways to alternatively target ABCB5 upregulation in CRC
ABCB5 upregulation is not directly targetable with pre-clinical or clinical compounds and the only ways to modify expression of this gene includes monoclonal antibody targeting or shRNA [45]. Although this is the case, we wanted to investigate whether it is possible to indirectly target ABCB5 by identifying druggable targets of the differentially expressed genes identified within our analysis (Table 1-2). We found three differentially expressed genes that are directly targetable with pre-clinical and/or clinical compounds: CD84, CDK1 and GHSR (Table 3). CDK inhibition is under clinical investigation in CRC and recently the combination of riviciclib and 5-FU was potent against CRC cells [46]. The relationship between 5-FU resistance mediated by ABCB5 upregulation and the consequential CDK1 upregulation highlights a vast and unexplored area of pre-clinical research. CD84 is targeted by the biological therapeutic etanercept. Normally etanercept is used to treat rheumatoid arthritis but has been explored in pre-clinical mouse colon cancer models showing anti-tumor effects and an enhancement of PDL1 and CTLA4 immunotherapy efficacy [47]. Identification of etanercept efficacy on ABCB5 positive population of CRCs would be a logical next step towards identifying the anti-tumor efficacy of this compound and further exploration into ways to overcome therapeutic resistance and enhancement of sensitivity. GSHR activity can be countered with the preliminary compound PF-5190457 but further investigatory work is needed to identify whether this compound harbors anti-cancer properties. Not only were some of the differentially expressed genes within this study targetable but also protein-network interactions involved with these differentially expressed genes were targetable with a variety of small molecule inhibitory compounds (Table 3).
Our research groups’ focus is mainly studying the impact of nuclear export protein 1 (XPO1) inhibition in gastrointestinal malignancies. Recently, the XPO1 inhibitor XPOVIO (selinexor) was FDA approved for use in multiple myeloma and diffuse large B cell lymphoma (DLBCL). Currently, selinexor is being investigated in CRC within a Phase I clinical trial [48]. Interestingly, we have found a connection between ABCB5 and XPO1 in CRC patient datasets as well as XPO1 and TROAP, a significantly upregulated gene caused by ABCB5 positivity (Figure 4A, Table 1). Trophinin Associated Protein (TROAP) is a microtubule and cytoskeleton re-arrangement protein, which pushes cells through the cell cycle [49]. After 24-hour treatment with selinexor, we found TROAP expression to be significantly downregulated in the HC-29 cell model, providing a simple example that selinexor could reverse the genetic consequences produced by ABCB5 upregulation (Figure 4B). Due to the vast underlying mechanisms in which ABCB5 upregulation alters cell cycle activity (Table 1-2) we found that 24-hour treatment with selinexor caused significant G1/S phase arrest in the HC-29 and HTC-116 colon cancer models (Figure 4C). These results suggest that selinexor has anti-cancer properties that can reverse some of the phenotypic changes associated with ABCB5 upregulation.
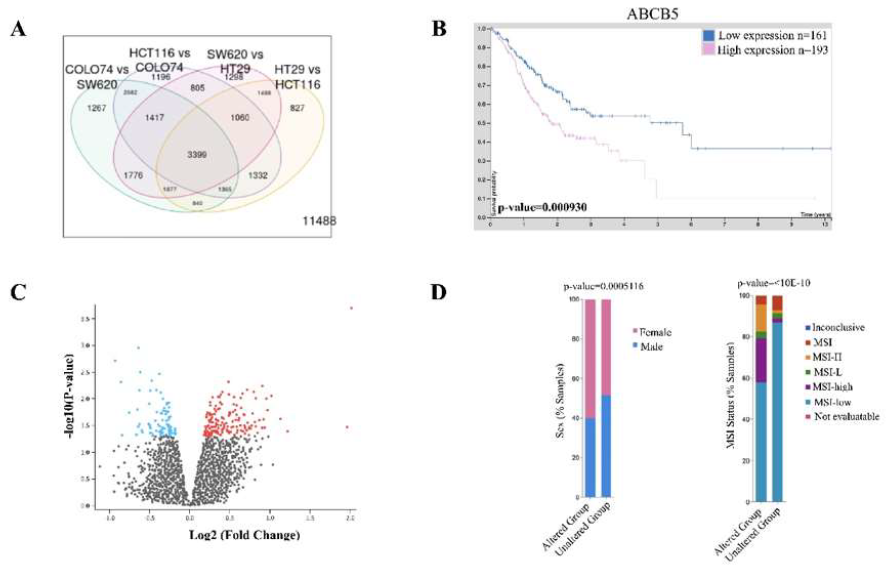
Figure 1: ABCB5 upregulation is prominent in CRC and produces widespread cellular changes. [A] Analysis of four colon cancer cell line models in GSE71670 provided by Geo2R database showing a variety of protein interactions common between cell lines. 3996 proteins were found to be common between all of the cell lines including ABCB5; [B] Human Protein atlas analysis of ABCB5 expression correlated to overall CRC patient survival. Statistics were provided by the software; [C] Volcano Plot obtained by GSE71670 dataset showing statistically and significantly changed proteins between ABCB5 positive and negative colon cancer cells of the four cell line models; [D] CBioPortal analysis of KIFC1, NDC80, KIF14 and UBE2C mutations in CRC patients stratifying for gender and MSI status. Statistics were provided by software.
|
Gene ID |
LogFC |
P-Value |
Cellular Function |
|
UBE2C |
3.97 |
0.00456 |
Ubiquitination |
|
ADGRF5 |
3.41 |
0.00995 |
G-protein coupled Receptor; Renal acid inhibitor |
|
MST1 |
3.29 |
0.0118 |
Immune System; Macrophage stimulating |
|
KIF14 |
3.29 |
0.0118 |
Cytoskeleton remodeling; microtubule motor protein |
|
SNORD1115-25 |
3.28 |
0.01193 |
Small Noncoding RNA |
|
DEFB126 |
3.13 |
0.01494 |
Polypeptides; inhibition of pathogenic invasion |
|
TNFSF8 |
3.02 |
0.01758 |
Immune system; tumor necrosis factor |
|
HJURP |
3.01 |
0.017771 |
DNA Damage repair; holiday junction protein |
|
NAG18 |
2.98 |
0.01869 |
Brain stimulating protein; function unknown |
|
FLJ35934 |
2.95 |
0.01934 |
Long Noncoding RNA |
|
MIRLET7C |
2.88 |
0.02162 |
MicroRNA |
|
WFDC6 |
2.86 |
0.02209 |
Serine Protease Inhibitor |
|
ANTXRL |
2.85 |
0.02264 |
Cellular Defense; Anthrax Toxin Defense Protein |
|
CENPL |
2.76 |
0.0258 |
Cellular Division; Centromere Protein |
|
EYS |
2.68 |
0.03284 |
Ocular Vision; Encodes rods and cones |
|
NDC80 |
2.6 |
0.03284 |
Cellular Division; kinetochore complex protein |
|
P2RY10 |
2.59 |
0.03333 |
G-protein coupled receptor |
|
CDCA8 |
2.58 |
0.03373 |
Cellular Division; Cell cycle arrest protein |
|
KIFC1 |
2.54 |
0.03621 |
Cellular division; mitotic spindle assembly |
|
KLKP1 |
2.52 |
0.03724 |
Androgen Signaling |
|
IGKC |
2.5 |
0.03835 |
Immune System; stimulating factor |
|
XAF1 |
2.48 |
0.03971 |
Inhibitor of Apoptosis protein |
|
TEX43 |
2.48 |
0.03972 |
Testis Expressed gene; Unknown function |
|
SERPINB4 |
2.45 |
0.04112 |
Protease inhibitor; inhibition of host immune response |
|
TROAP |
2.44 |
0.04182 |
Cell Adhesion; trophin molecule |
|
KIF18B |
2.44 |
0.04214 |
Cytoskeleton; Microtubule structure |
|
CD84 |
2.43 |
0.04256 |
Cell surface immunoreceptor |
|
PAGE2 |
2.43 |
0.04272 |
P-antigen; Unknown function |
|
C14orf178 |
2.42 |
0.0433 |
Unknown |
|
CDK1 |
2.41 |
0.04373 |
Cellular Division; Activator of Cell cycle |
|
CCL4L1///CCL4L2///CCL4///CCL4L1 |
2.41 |
0.04402 |
Chemokine; stimulator of immune system activity |
|
STATH |
2.41 |
0.04432 |
Inhibitor of calcium precipitation, phosphate buffer |
|
GHSR |
2.39 |
0.04559 |
G-protein coupled receptor; stimulates growth hormone |
|
CCNF |
2.35 |
0.04803 |
Cellular Division; Activator of Cell cycle |
|
HIST1H4D |
2.34 |
0.04929 |
Epigenetic modifications; histone complex |
|
ACKR4 |
2.33 |
0.04973 |
Chemokine Receptor; CCL21 regulation |
Table 1: Comprehensive Table of Upregulated Genes found in GSE71670 Dataset. Cellular Function was determined from KEGG pathway analysis.
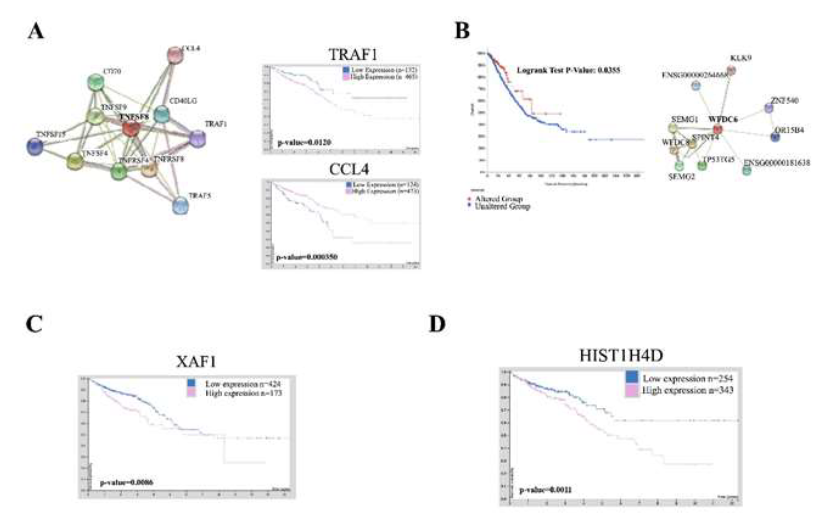
Figure 2: Upregulated genes caused by ABCB5 upregulation are predictive of disease in CRC. [A] STRING database analysis of TNFSF8 showing protein-protein interactions. Human protein analysis survival curves of CRC patients with TRAF1 and CCL4 expression. The software provided statistics; [B] CBioPortal analysis of CRC patients with mutations in WFDC6 and other interacting proteins (provided by STRING database). Statistics were provided by the software; [C] Human Protein atlas showing survival of CRC patients of all stages with XAF1 expression. Statistics were provided by software; [D] Human Protein Atlas showing survival of CRC patients overall with HIST1H4D expression. Statistics were provided by software.
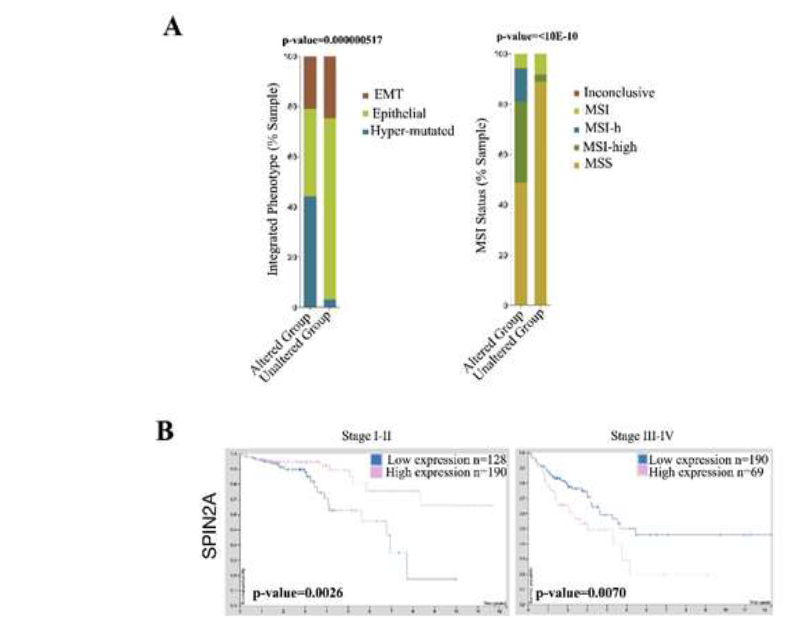
Figure 3: Downregulated genes caused by ABCB5 upregulation are predictive of disease in CRC. [A] CBioPortal Analysis of all olfactory receptor family members, which interact, with OR4A15. Statistics provided by software. [B] Human Protein analysis of CRC patient survival with SPIN2A expression subcategorized by stage. The software provided statistics.
|
Gene ID |
LogFC |
P-Value |
Cellular Function |
|
OR5M3 |
-3.45 |
0.00937 |
G-protein coupled receptor; olfactory signaling |
|
PTTG3P |
-3.3 |
0.01156 |
Cell cycle; long-noncoding RNA |
|
LIMD1-AS1 |
-3.24 |
0.01265 |
Long-noncoding RNA; tumor suppressor |
|
GPR151 |
-2.95 |
0.01936 |
G-protein coupled receptor; pH sensing |
|
LOC102722437 |
-2.87 |
0.02201 |
Unknown |
|
OR5AK2 |
-2.82 |
0.02354 |
G-protein coupled receptor; olfactory signaling |
|
FAM167A-AS1 |
-2.8 |
0.02423 |
Long-noncoding RNA; unknown |
|
ACRV1 |
-2.79 |
0.0246 |
Differentiation Protein; testis specific |
|
OR4A15 |
-2.77 |
0.02534 |
G-protein coupled receptor; olfactory signaling |
|
C12orf50 |
-2.73 |
0.02719 |
Unknown |
|
HIGD1B |
-2.72 |
0.02727 |
Hypoxia inducing gene |
|
SCARNA4 |
-2.61 |
0.0323 |
snoRNA |
|
SPIN2A |
-2.59 |
0.03369 |
Cell cycle; inhibitor of cell cycle progression |
|
FBXO47 |
-2.58 |
0.03375 |
F-box protein; meiosis |
|
FAM205BP |
-2.58 |
0.0339 |
Small-noncoding RNA |
|
SNORD117 |
-2.47 |
0.03996 |
snoRNA |
|
SNORD63 |
-2.47 |
0.04035 |
snoRNA |
|
OR6C3 |
-2.46 |
0.04098 |
G-protein coupled receptor; olfactory signaling |
Table 2: Comprehensive Table of Downregulated Genes found in GSE71670 Dataset. Cellular Function was determined from KEGG pathway analysis.
|
Gene ID |
Drug/Targetable Gene |
|
CD84 |
Etanercept |
|
CDK1 |
Riviciclib, AG-24322, Dinaciclib, Boheime, RG-547, Milciclib |
|
GHSR |
PF-5190457 |
|
UBE2C |
ACE (cilazapril, spirapril, liniospril, quinapril, benazepril, moexipril), ACE2 (ORE-1001, GSK-2586881, Lisinopril, captopril), |
|
ADGRF5 |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), , HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRM2 (JNJ-40411813, LY2969822, LY404039), GRIK4 (dysiherbaine, tezampanel, selurampanel), ERBB4 (afatinib, tucatinib, allitinib, poziotinib, dacomitinib), PLK2 (onvansertib, BI-2536, Wortmannin), CAMK2D (gefitinib, sotrastaurin, LY-2090314), ABL2 (dasatinib, ponatinib, pemetinib, Imatinib, tozasertib), CAMK2A (curcumin, AZD-1080, Sotrastaurin, Cenisertib), DYRK2 (leucettamine B, Harmine), MAP3K12 (CEP-1347) |
|
MST1 |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), , HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRM2 (JNJ-40411813, LY2969822, LY404039), GRIK4 (dysiherbaine, tezampanel, selurampanel), ERBB4 (afatinib, tucatinib, allitinib, poziotinib, dacomitinib), PLK2 (onvansertib, BI-2536, Wortmannin), CAMK2D (gefitinib, sotrastaurin, LY-2090314), ABL2 (dasatinib, ponatinib, pemetinib, Imatinib, tozasertib), CAMK2A (curcumin, AZD-1080, Sotrastaurin, Cenisertib), DYRK2 (leucettamine B, Harmine), MAP3K12 (CEP-1347) |
|
DEFB126 |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), , HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRM2 (JNJ-40411813, LY2969822, LY404039), GRIK4 (dysiherbaine, tezampanel, selurampanel), ERBB4 (afatinib, tucatinib, allitinib, poziotinib, dacomitinib), PLK2 (onvansertib, BI-2536, Wortmannin), CAMK2D (gefitinib, sotrastaurin, LY-2090314), ABL2 (dasatinib, ponatinib, pemetinib, Imatinib, tozasertib), CAMK2A (curcumin, AZD-1080, Sotrastaurin, Cenisertib), DYRK2 (leucettamine B, Harmine), MAP3K12 (CEP-1347) |
|
WFDC6 |
SERPIND1 (sulodexide), SERPINE1 (tiplasinin, aleplasinin, urokinase, diaplasinin, dalteparin), AGT (quinapril, imidapril, benazepril, mestranol, adriamycin, enalapril), |
|
ANTXRL |
ABCB1 (voacamine, zosuqidar, tariquidar), ADAM10 (aderbasib, marimastat, ilomastat), ADAM17 (chembl489100, aderbasib, apratastat, marimastat), CD36 (ABT-510), MST1R (MK-8033, Glesatinib, MGCD-265, BMS-777607), NT5E (oleclumab, tinidazole, vesnarinone, capsaicin, reserpine, lovastatin) |
|
P2RY10 |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), , HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRM2 (JNJ-40411813, LY2969822, LY404039), GRIK4 (dysiherbaine, tezampanel, selurampanel, topiramate), GRM3 (LY404039, Spaglumic acid, LY2969822, Risperidone), HRH4 (toreforant, PF-03893787, UR-63325, JNJ-7777120), GPR84 (GLPG-1205, Setogepram), PTAFR (rupatadine, bepafant, israpafant, Minopafant, CV-6209) TSHR (thyrotropin alpha, coumarin-3-Carboxylic acid, CHEMBL584905, CEMBL249190) |
|
CDCA8 |
ERBB4 (afatinib, tucatinib, allitinib, poziotinib, dacomitinib), PLK2, CAMK2D (gefitinib, sotrastaurin, LY-2090314), ABL2 (Dasatinib, Ponatinib, Pemetinib, Imatinib), CAMK2A (AZD-1080, Curcumin, CYC-116), DYRK2 (Leucettamine B, Harmine), MAP3K12 (CEP-1347) |
|
KLKP1 |
XPNPEP1 (Tosedostat), HGF (Rilotumumab, ficlatuzumab, MP-0250, Wortmannin, Cabozantinib), METAP2 (Beloranib, PPI-2458, Tosedosat), MMP7 (Marimastat, Doxycycline) |
|
IGKC |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRIK4 (dysiherbaine, tezampanel, selurampanel, topiramate), GRM3 (LY404039, Spaglumic acid, LY2969822, Risperidone), SLC5A1 (Sotagliflozin, GSK-1614235, Canagliflozin, Licogliflozin), SLC29A4 (Quinine, Cimetidine, Metformin, Fluoxetine, Verapamil), SLC5A3 (Phlorizin), SLC36A1 (Oxitriptan, Tryptophan, Serotonin) |
|
SERPINB4 |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRIK4 (dysiherbaine, tezampanel, selurampanel, topiramate), GRM2 (JNJ-40411813, LY2969822, LY404039), SERPIND1 (Sulodexide), SERPINE1 (Tiplasinin, Aleplasinin, Cetrorelix, Dalteparin), AGT (Quinapril, CYT006-ANGQB, Imidapril, Lisinopril, Atenolol, Enalapril), ACE (Cilazapril, Lisinopril, Spirapril), ACE2 (ORE-1001, Lisinopril, GSK-2586881, Captopril) |
|
ACKR4 |
HRH4 (Toreforant , PF-03893787, UR-63325, JNJ-7777120), GPR84 (GLPG-1205, Setogepram), PTAFR (rupatadine, Bepafant, Israpafant, CV-6209), TSHR (Thyrotropin Alpha, CEMBL584905, CEMBL249190, CEMBL578276), PTGS1 (Suprofen, Salsalate, Bromfenac, Oxaprozin, Ketoprofen), HCN3 (Zatebradine, cilobradine, Ivabradine), CCR2 (AZD2423, CCX140, Plozalizumab, MK-0812), GRM3 (LY404039, LY2969822, Risperidone), GRIK4 (Dysiherbaine, Haloperidol, Topiromate, Texampanel), SLC5A1 (Gotagliflozin, Canagliflozin, Remogliflozin, Ipragliflozin, Empagliflozin), SLC29A4 (Quinine, Cimetidine, Desipraminine, Quinidine, Metformin), SLC5A3 (Phlorizin), SLC36A1 (Oxitriptan, Tryptophan, Serotonin) |
|
OR5M3 |
PTGS1 (suprofen, salsalate, tolmetin, ketoprofen, ginger, piroxicam, naproxen, curcumin), , HCN3 (zaterbradine, cilobradine, ivabradine), CCR2 (AZD2423, CCX140, plozalizumab, MK-0812), GRM2 (JNJ-40411813, LY2969822, LY404039), GRIK4 (dysiherbaine, tezampanel, selurampanel), ERBB4 (afatinib, tucatinib, allitinib, poziotinib, dacomitinib), PLK2 (onvansertib, BI-2536, Wortmannin), CAMK2D (gefitinib, sotrastaurin, LY-2090314), ABL2 (dasatinib, ponatinib, pemetinib, Imatinib, tozasertib), CAMK2A (curcumin, AZD-1080, Sotrastaurin, Cenisertib), DYRK2 (leucettamine B, Harmine), MAP3K12 (CEP-1347) |
Table 3: Comprehensive List of directly or indirectly druggable genes found within the GSE71670 dataset. Gene-drug interactions were obtained from DGIdb and not all possible drug interactions are listed within the table.
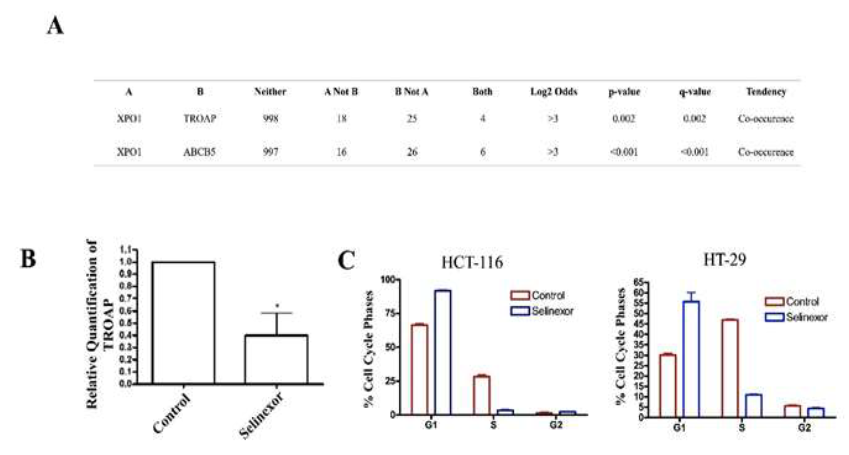
Figure 4: Selinexor Interacts with Genes Altered by ABCB5. [A] CBioPortal Analysis of TROAP and ABCB5 in relation to XPO1 in CRC cohort. Statistics provided by software; [B] RT-qPCR analysis of TROAP expression after 24-hour treatment with Selinexor (500 nM); [C] Cell cycle analysis of HCT-116 and HT-29 cell line models after 24-hour treatment with Selinexor; 400 nM and 500 nM respectively.
4. Conclusion
Colorectal Cancer (CRC) is a complex and deadly disease. We have assessed a profile of genetic targets altered by ABCB5 upregulation in CRC. Regardless of the gene found, whether a focus of pre-clinical research or not, there has been no previous concrete evidence linking the expression of these perturbed proteins to ABCB5 upregulation. Further, there has been no evidence linking perturbation of small-noncoding RNAs to ABCB5 upregulation. Small-noncoding RNAs were identified as drivers of CRC pathogenesis, progression and disease and a better understanding of the native function of these small noncoding RNAs and the mechanisms in which they are altered by ABCB5 may be useful towards future prognostic or predictive biomarker studies [50]. Interestingly we have previously linked XPO1 upregulation to the perturbation of a subset of cancer specific small-noncoding RNAs [51] and further investigation into this axis within CRC is necessary. Not only have we identified genetic aberrations caused by ABCB5 but also have provided predicted in-direct druggable targets that can be explored in future studies.
Although selinexor was not identified within our analysis as a direct target, we have found that treatment of colon cancer cells with this compound was effective in reducing aberrations caused by ABCB5 positivity. This finding correlates with previous studies suggesting that selinexor can target the stem cell population as well as differentiated malignant cells [52]. Identifying ABCB5 status within CRC patients may allow for the identification of an ideal patient population that would benefit from XPO1 inhibitory therapy or may lay the groundwork for future biomarker studies. The result of this study includes (1) aiding in future drug discoveries and (2) identifying novel therapies that can be tested pre-clinically in CRC models. Targeting ABCB5 is a promising strategy for targeting CRC growth and progression and it is clear that further preclinical investigation is needed.
Conflict of Interest Statement
Conflicts that are not related to this study. ASA received funding and speaker fees from Karyopharm Therapeutics for independent projects. ASA received funding from Janssen, Rhizen and EISAI for independent projects. ASA serves as a consultant for GLG and Guidelpoint.
References
- Colorectal Cancer Facts & Figures: Facts About Colon Cancer. American Cancer Society (2021).
- Cancer of the Colon and Rectum - Cancer Stat Facts. SEER (2021).
- Colon Cancer. Mayo Clinic, Mayo Foundation for Medical Education and Research (2020).
- Hammond, William A, Mody Kl. Pharmacologic Resistance in Colorectal Cancer: a Review. Therapeutic Advances in Medical Oncology, SAGE Publications 8 (2016): 57-84.
- Yuan-Hong Xie, Ying-Xuan Chen, Jing-Yuan Fang. Comprehensive Review of Targeted Therapy for Colorectal Cancer. Nature News, Nature Publishing Group 22 (2020).
- Voutsadakis, Ioannis A. The Ubiquitin-Proteasome System in Colorectal Cancer. Biochimica Et Biophysica Acta (BBA) - Molecular Basis of Disease, Elsevier (2018).
- Drugs Approved for Colon and Rectal Cancer. National Cancer Institute (2020)
- Van der Jeught K, Xu HC, Li YJ, et al. Drug resistance and new therapies in colon cancer. World J Gastroenterol 24 (2018): 3834-3848.
- Kugimiya N, Nishimoto A, Hosoyama T, et al. The c-MYC-ABCB5 axis plays a pivotal role in 5-fluorouracil resistance in human colon cancer cells. J Cell Mol Med 19 (2015): 1569-1581.
- Lin JY, Zhang M, Schatton T, et al. Genetically determined ABCB5 functionality correlates with pigmentation phenotype and melanoma risk. Biochem Biophys Res Commun 436 (2013): 536-542.
- Kondo S, Hongama K, Hanaya K, et al. Upregulation of cellular glutathione levels in human ABCB5 and murine Abcb5-transfected cells. BMC Pharmacology and Toxicology 16 (2015).
- Lehne G, Grasmo-Wendler UH, Berner JM, et al. Upregulation of stem cell genes in multidrug resistant K562 leukemia cells. Leukemia Research 33 (2009): 1379-1385.
- Guo G, Grimmig T, Gonzalez G, et al. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J Biol Chem 293 (2018): 11166-11178.
- Jianjiong Gao, Bülent Arman Aksoy, Ugur Dogrusoz, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6 (2013): 11.
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 (2002): 207-210.
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41 (2013): D991-D995.
- Uhlén M, Fagerberg L, Hallstrom BM, et al. Tissue-based map of the human proteome. Science (2015).
- Freshour S, Kiwala S, Cotto KC, et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research (2020).
- https://www.proteinatlas.org/ENSG00000132530-XAF1/pathology/colorectal+cancer
- https://www.proteinatlas.org/ENSG00000147059-SPIN2A/pathology/colorectal+cancer
- https://www.proteinatlas.org/ENSG00000106952-TNFSF8
- https://www.proteinatlas.org/ENSG00000277157-HIST1H4D/pathology/colorectal+cancer
- https://www.proteinatlas.org/ENSG00000004846-ABCB5/pathology/colorectal+cancer
- Wang ZZ, Yang, Jiang BH, et al. KIF14 promotes cell proliferation via activation of AKT and is directly targeted by miR-20 in colorectal cancer. International Journal of Oncology (2018): 1939-1952.
- Sishtla K, Pitt N, Shadmand M, et al. Observations on spontaneous tumor formation in mice over expressing mitotic kinesin KIF14. Scientific Reports 8 (2018).
- Akabane S, Oue N, Sekino Y, et al. KIFC1 regulates ZWINT to promote tumor progression and spheroid formation in colorectal cancer. Pathol Int (2021).
- Yan X, Huang L, Liu L, et al. Nuclear division cycle 80 promotes malignant progression and predicts clinical outcome in colorectal cancer. Cancer Med 7 (2018): 420-432.
- Thiru P, Kern DM, McKinley KL, et al. Kinetochore genes are coordinately up-regulated in human tumors as part of FoxM1-related division program. Mol Biol Cell 25 (2014): 1983-1994.
- Bavi P, Uddin S, Ahmed M, et al. Bortezomib Stabilizes Mitotic Cyclins and Prevents Cell Cycle Progression via Inhibition of UBE2C in Colorectal Carcinoma. Am J Pathol 178 (2011): 2109-2120.
- Fujita T, Ikeda H, Taira N, et al. Overexpression of UbcH10 alternates the cell cycle profile and accelerate the tumor proliferation in colon cancer. BMC Cancer 9 (2009): 87-10.
- Zhan Z, Li L.Y. TNFSF15 Modulates Neovascularization and Inflammation. Cancer Microenviron 5 (2012): 237-247.
- Tuomisto AE, Makinen MJ, Varyrynen JP. Systemic inflammation in colorectal cancer: Underlying factors, effects and prognostic significancer. World J Gastroenterol 25 (31): 4383-4404.
- Duffy MJ. Do proteases play a role in cancer invasion and metastasis?. Eur J Cancer Clin Oncol 23 (1987): 583-589.
- Jacquelot N, Yamazaki T, Roberti MP, et al. Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell research (2019).
- Silke J, Meier P. Inhibitor of Apoptosis (IAP) Proteins-Modulators of Cell Death and Inflammation. Cold Spring Harb Perspect Biol 5 (2013): a008730.
- Xie W, Zhang J, Zhong P, et al. Expression and potential prognostic value of histone family gene signature in breast cancer. Exp Ther Med 18 (2019): 4893-4903.
- Al-Temaimi RA, Tan TZ, Marafie MJ, et al. Identification of 42 Genes Linked to Stage II colorectal Cancer Metastatic Relapse. Int J Mol Sci 17 (2016): 598.
- Gad AA, Balenga N. The Emerging Role of Adhesion GPCRs in Cancer. ACS Publications (2020).
- Lea Weber, Klaudia Al-Refae, Juliane Ebbert, et al. Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis. PLOS ONE 12 (2017): e0172491.
- Fletcher BS, Dragstedt C, Notterpek L, et al. Functional cloning of SPIN-2, a nuclear anti-apoptotic proteins with roles in cells cycle progression. Leukemia 16 (2002): 1507-1518.
- Pan J, Tang Y, Liu S, et al. LIMD1-AS1 suppressed non-small cell lung cancer through stabilizing LIMD1 mRNA via hnRNP U. Cancer Med 9 (2020): 3829-3839.
- Shang X, Song X, Wang K, et al. SNORD63 and SNORD96A as the non-invasive diagnostic biomarkers for clear cell renal cell carcinoma. Cancer Cell International 56 (2021).
- Gunaldi M, Kocoglu H, Okuturlar Y, et al. Heat shock protein 70 is a useful marker for predicting colorectal cancer. J BUON 20 (2015): 1464-1470.
- Peelman LJ, Chardon P, Nunes M, et al. The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family. Genomics 26 (1995): 210-218.
- Diaz A, Leon K. Therapeutic Approaches to Target Cancer Stem Cells. Cancers (Basel) 3 (2011): 3331-3351.
- Li PM, Lee HM, Huang C, et al. Synergistic Antiproliferative Effect of Ribociclib (LEE011) and 5-Fluorouracil on Human Colorectal Cancer. Anticancer Research 40 (2020): 6265-6271.
- Montfort A, Colacios C, Levade T, et al. The TNF Paradox in Cancer Progression and Immunotherapy. Frontiers in Immunology 10 (2019): 1818.
- Mau-Soerensen M, Razak ARA, Mahipal A, et al. Safety and anti-tumor activity of selinexor (KPT-330), a first-in-class, oral XPO1 selective inhibitor of nuclear export: A phase I study expanded with colon cancer cohort. Journal of Clinical Oncology 32 (2014).
- Li K, Zhang R, Wei M, et al. TROAP Promotes Breast Cancer Proliferation and Metastasis. BioMed Research International (2019): 6140951.
- Wang J, Wang X, Song YX, et al. Circulating Noncoding RNAs have a promising future as Novel Biomarkers for Colorectal Cancer. Disease Markers (2018).
- Sexton R, Mahdi Z, Chaudhury R, et al. Targeting Nuclear Exporter Protein XPO1/CRM1 in Gastric Cancer. International Journal of Molecular Sciences (2019).
- Than H, Pomicter AD, Yan D, et al. Coordinated inhibiton of nuclear export and BCR-Abl1 selectively targets chronic myeloid leukemia stem cells. Leukemia 34 (2020): 1679-1683.
