Genetic characterization of cystic pulmonary echinococcosis in Sudan as determined by mitochondrial DNA sequences
Article Information
Sara S Abdalla1,2, Mohamed E Ahmed2,3, Alsmawal Awad Elimam5, Mawahib H Eldigail4, Imadeldin E Aradaib2,4, Martin P Grobusch2,6
1Department of Medical Parasitology and Entomology, Faculty of Medical laboratory Sciences, AlNeelain University, Khartoum, Sudan
2Medical Research Center, Zamzam University College, Khartoum, Sudan
3Department of Surgery, Faculty of Medicine, AlNeelain University, Khartoum, Sudan
4Vetrinery Molecular Biology Laboratory, Khartoum University, Khartoum, Sudan
5Department of Histopathology and cytology, Faculty of Medical Laboratory Sciences, AlNeelainUniversity, Khartoum, Sudan
6Center for Tropical Medicine and Travel Medicine, Department of Infectious Diseases, Amsterdam University Medical Centres, location AMC, Amsterdam Infection & Immunity, Amsterdam Public Health, University of Amsterdam, The Netherlands
*Corresponding author: Mohamed E Ahmed, Zamzam Medical Research center, Zamzam University College and Department of surgery, Faculty of Medicine, AlNeelain University, Khartoum, Sudan
Received: 11 June 2021; Accepted: 18 June 2021; Published: 16 July 2021
Citation: Sara S Abdalla, Mohamed E Ahmed, Alsmawal Awad Elimam, Mawahib H Eldigail, Imadeldin E Aradaib, Martin P Grobusch. Genetic characterization of cystic pulmonary echinococcosis in Sudan as determined by mitochondrial DNA sequences. Journal of Surgery and Research 4 (2021): 353-362.
Share at FacebookAbstract
Background: Cystic echinococcosis (CE) is a zoonotic parasitic disease caused by the larval stages of the cestode Echinococcus granulosus. It is a significant public health problem with high endemicity in east and central Africa including Sudan. Worldwide, pulmonary echinococcosis cyst is a significant problem medically, socially, and economically. Until now ten genetic variants, or genotypes designated as (G1-G10), are distributed worldwide based on genetic diversity. In this study we aimed to investigate molecular diversity of Echinococcus granulosus isolates collected from human clinical samples removed surgically from lung using mitochondrial gene nad1 in Sudan.
Methods: Seven human echinococcosis cysts originated from lung were collected through surgery from ElShaab Teaching Hospital in Khartoum state during 2016-2018. The fertility of cysts was detected microscopically by examination of the cysts fluid in which the protoscoleces were found. Protoscoleces were removed from each cyst and their total genomic DNAs were extracted. PCR was performed to amplify fragments of 530 base pair (bp) forNADH dehydrogenase subunit 1(NADH-1) gene. The amplified PCR products were purified and subjected to direct sequencing for subsequent construction of phylogenetic tree and network analysis.
Results: The identity of the PCR products were confirmed as NADH1 nucleotide sequences using the Basic Local Alignment Search Tool (BLAST) of NCBI (National Center for Biotechnology Information, Bethesda, MD).The phylogenetic tree revealed the presence of Echinococcus canadenesis genotype 6 (G6) in all cysts (100%).
Conclusions: According to the results of the previous and present observations which has been performed by PCR only and PCR followed by sequencing respectively, it can be
Keywords
Echinococcosis, Pulmonary, PCR, DNA sequencing, Phylogony, Sudan
Echinococcosis articles; Pulmonary articles; PCR articles; DNA sequencing articles; Phylogony articles; Sudan articles
Echinococcosis articles Echinococcosis Research articles Echinococcosis review articles Echinococcosis PubMed articles Echinococcosis PubMed Central articles Echinococcosis 2023 articles Echinococcosis 2024 articles Echinococcosis Scopus articles Echinococcosis impact factor journals Echinococcosis Scopus journals Echinococcosis PubMed journals Echinococcosis medical journals Echinococcosis free journals Echinococcosis best journals Echinococcosis top journals Echinococcosis free medical journals Echinococcosis famous journals Echinococcosis Google Scholar indexed journals Pulmonary articles Pulmonary Research articles Pulmonary review articles Pulmonary PubMed articles Pulmonary PubMed Central articles Pulmonary 2023 articles Pulmonary 2024 articles Pulmonary Scopus articles Pulmonary impact factor journals Pulmonary Scopus journals Pulmonary PubMed journals Pulmonary medical journals Pulmonary free journals Pulmonary best journals Pulmonary top journals Pulmonary free medical journals Pulmonary famous journals Pulmonary Google Scholar indexed journals PCR articles PCR Research articles PCR review articles PCR PubMed articles PCR PubMed Central articles PCR 2023 articles PCR 2024 articles PCR Scopus articles PCR impact factor journals PCR Scopus journals PCR PubMed journals PCR medical journals PCR free journals PCR best journals PCR top journals PCR free medical journals PCR famous journals PCR Google Scholar indexed journals DNA sequencing articles DNA sequencing Research articles DNA sequencing review articles DNA sequencing PubMed articles DNA sequencing PubMed Central articles DNA sequencing 2023 articles DNA sequencing 2024 articles DNA sequencing Scopus articles DNA sequencing impact factor journals DNA sequencing Scopus journals DNA sequencing PubMed journals DNA sequencing medical journals DNA sequencing free journals DNA sequencing best journals DNA sequencing top journals DNA sequencing free medical journals DNA sequencing famous journals DNA sequencing Google Scholar indexed journals Phylogony articles Phylogony Research articles Phylogony review articles Phylogony PubMed articles Phylogony PubMed Central articles Phylogony 2023 articles Phylogony 2024 articles Phylogony Scopus articles Phylogony impact factor journals Phylogony Scopus journals Phylogony PubMed journals Phylogony medical journals Phylogony free journals Phylogony best journals Phylogony top journals Phylogony free medical journals Phylogony famous journals Phylogony Google Scholar indexed journals circulating genetic variants articles circulating genetic variants Research articles circulating genetic variants review articles circulating genetic variants PubMed articles circulating genetic variants PubMed Central articles circulating genetic variants 2023 articles circulating genetic variants 2024 articles circulating genetic variants Scopus articles circulating genetic variants impact factor journals circulating genetic variants Scopus journals circulating genetic variants PubMed journals circulating genetic variants medical journals circulating genetic variants free journals circulating genetic variants best journals circulating genetic variants top journals circulating genetic variants free medical journals circulating genetic variants famous journals circulating genetic variants Google Scholar indexed journals aspirated fluid samples articles aspirated fluid samples Research articles aspirated fluid samples review articles aspirated fluid samples PubMed articles aspirated fluid samples PubMed Central articles aspirated fluid samples 2023 articles aspirated fluid samples 2024 articles aspirated fluid samples Scopus articles aspirated fluid samples impact factor journals aspirated fluid samples Scopus journals aspirated fluid samples PubMed journals aspirated fluid samples medical journals aspirated fluid samples free journals aspirated fluid samples best journals aspirated fluid samples top journals aspirated fluid samples free medical journals aspirated fluid samples famous journals aspirated fluid samples Google Scholar indexed journals
Article Details
1. Introduction
Cystic echinococcosis, caused by the metacestode of Echinococcus granulosus, is one of the most common zoonotic diseases associated with huge economic losses and great public health significance worldwide [1]. Dog and other carnivores that harbor the adult cestode in their small intestine are the definitive hosts for the parasite, while a wide range of mammalian species including domestic animals and man act as intermediate hosts [2]. The metacestode stage of E. granulosus inhabits the liver, lungs, and other internal organs of livestock and humans after oral uptake of the eggs, which are produced by adult worms in the canine small intestine [3,4]. Humans can be accidently infected by ingesting its eggs, which results in human cystic echinococcosis (CE) or hydatidosis [5]. CE may develop silently over years and even decades until it surfaces with clinical signs or symptoms. Clinical symptoms are mainly related to the localization, size and number of cysts. However, a recent study described that there seemed to be a relationship between the genotypes and the size of hydatid cysts, in which all the patients infected with G7 genotype showed smaller liver cysts than those infected with G1 genotype. CE is an important and sometimes life-threatening disease, which mostly affects lungs and liver, but other organ scan be also affected [6]. Fatal cases of human CE have been reported with inoperable CE cysts of the brain and anaphylactic shock caused by rupture of liver hydatid cysts [7]. In the Sudan, many reports of cystic echinococcosis have been described in humans and animals [8]. In addition to its importance as a major public health problem in the country, CE is also considered as one of the major causes of condemnation of sheep carcasses during meat inspection. Different genotypes of E.granulosus have been identified from a variety of hosts worldwide. Until now, 10 genotypes(G1-G10) has been described [9]. The genotypes are different in some criteria such as pathogenicity, host specificity, pattern of life cycle, transmission dynamics and developmental rates, human infectivity and response to chemotherapeutic drugs [10]. Of the ten genotypes of EG, the sheep(G1), the cattle (G5) and the camel (G6) strains were reported in humans and livestock in the Sudan [11,12]. The epidemiological situation in Sinnar area, Blue Nile state is characterized by intense transmission of Echinococcus canadensis(G6), thereby closely resembling the situation in most other regions of Sudan [13]. This study aimed to investigate genotypes of the echinococcosis cysts isolated from echinococcosis patients from different parts of Sudan. Echinococcosis cysts were defined by genetic studies and subsequent phylogenetic analysis. The molecular characterization was made possible by targeting fragments of the mitochondrial NADH 1 to define the circulating genetic variants in the different part of Sudan.
2. Materials and Methods
2.1 Collection of samples
A total of 7 Echinococcus cysts were collected from 7 patients suffering from pulmonary CE. All patients came from different parts of Sudan and were operated surgically at Al-Shaab hospital. The cysts were preserved separately in 70% ethanol and formalin.
2.2 Microscopic examination
Cysts or cyst material were examined microscopically toconfirm whether or not they are hydatid cysts. Fertility of the collected Echinococcus cysts was assured by detection of protoscolices in aspirated fluid samples.
2.3 Extraction of genome DNA
Genomic DNA was extracted from sediment positive for protoscoleces using GF-1 Blood DNA Extraction Kit (Vivantis) according to manufacturer’s instructions. Briefly, 200 μl of the suspended aspirate, 20 μl of proteinase K stock solution, and 200 μl of lysing buffer were pipetted into 1.5 ml eppendorf tube. The mixture was incubated at 65°C for 10 min before the addition of 200 μl of absolute alcohol and mixing by vortexing. The mixture was then transferred to the spin column placed in a clean 2 ml collection tube and centrifuged at 5000 RPM in MiniSpin centrifuge (Eppendorf, Wesseling- Berzdorf, Germany) for 1 min at room temperature. The spin column was washed twice with 500 μl of the washing buffersand centrifuge at 5000 RPM for 1 min and the DNA was eluted in 100µl of buffer and was stored at 4 ° c until further use in PCR analysis.
2.3.1 Selection of primers: The primers were selected from mitochondrial NADH dehydrogenase subunit 1 (NADH 1) gene. The NADH 1 primers used in this study were basically described previously (Bowles J and McManus DP, 1993). For the first amplification step, a pair of outerprimers JBl1: 5' AGATTCGTAAGGGGCCTAATA 3' andJB12: 5' ACCACTAACTAATTCACTTTC 3' were used toamplify a 530 bp PCR product from EG isolates. A pair of internal sequencing primers JB11.5: 5' TTATGGTAGATATTATAG 3' and JB12.5: 5' CACACACATAAAACAAGC 3' designed to conserved segments of the EG NADH1 sequences, were used to generate a 471 bp PCR product
2.4 Polymerase chain reaction (PCR)
A stock buffered solution containing 150 μl 10x PCR buffer, 100 μl of 25 mM MgCl2, 12.5 μl of each dATP, dTTP, dGTP and dCTP at a concentration of 10 mM was prepared in 1.5 ml eppendorf tube. The primers were used at a concentration of 20 pg/μl, and double distilled water was added to bring the volume of the stock buffer solution to 1.5 ml. Each 0.5 ml PCR reaction tube contained 2 μl of the primers, 1 μl (5.0 U) of Taq DNA polymerase (Vivants), 5.0 μl of the target DNA and 42 μl of the stock buffered solution. For nested PCR, 2 μl of the primary PCR product was used as DNA template. The thermal cycling profiles were as follows: a 2 min initial incubation at 95°C, followed by40 cycles of 95°C for 1 min, 55°C for 30 sec and 72°C for 45 sec, and a final incubation at 72°C for 10 min. Thermal profiles were performed on a Techne TC-412 thermal cycler (Techne, Staffordshire, UK). Following amplification, 15 μl from each PCR containing amplified products wereloaded onto gels of 1.0% agarose and electrophoresed for 1 h. The gels were stained with ethidium bromide and the PCR products were easily identified following visualization under UV light.
2.5 Sequence analysis and construction of phylogenetic tree
The sequences obtained from the PCR products were sent for sequencing to commercial company (Macrogen, Seoul, Korea). Resulted sequences were edited and aligned using BioEdit software (Ibis Biosciences, Carlsbad, CA, USA). The Basic Local Alignment Search Tool (BLAST) of NCBI (National Centre for Biotechnology Information, Bethesda, MD, USA) was used to confirm the identity of the generated sequences in relation to GenBank nucleotide database. The sequences then aligned with the corresponding region of NADH1 subunit gene of known genotypes from other countries. All sequences were submitted to the GenBank (accession numbers MW660881 to MW660886). The phylogenetic tree was constructed using Neighbor joining tree implemented in MEGA software version 7.
3. Results
3.1 Polymerase chain reaction (PCR)
The PCR-based assay with primers specific for NADH-1 yielded amplification products from of all of the seven hydatid cysts obtained from patients. For NADH1, the outer pair of primers produced a primary 530 bp PCR product and the nested primers produced a 471 bp PCR product (figure 1). The PCR products submitted for sequencing to a commercial company (Macrogen, Seoul, Korea).
3.2 Sequence analysis and phylogenetic relationship
The sequences obtained from the PCR products were found to aligned and compared with corresponding regions for NADH1 gene in the GenBank confirming the cysts to contain the EG complex, using BioEdit and also the BLAST program of GenBank. All sequences were submitted to the GenBank (accession nos. MW660881 to MW660886). Genotype identifications were in complete agreement for all isolates. Blast search of the obtained sequences derived from human isolates indicated the occurrence of G6 genotype (E. canadenesis) in all isolates. Presence of nucleotide substitution at position 16282(A to C) was detected among 2 strains with G6 genotype when compare to reference strain NC_044548.1 (complete genome), also found other substitution at position 7579 (G to C) in that same 2 strains when compared to reference strain NC_038227.1(G6 strain) (Figure 2). Among all the strains with G6 genotype, substitution at position 16132 (C to G) was observed in compare to reference strain NC_044548.1 (complete genome) and NC_038227.1(G6 strain) (Figure 3). The phylogenetic analysis of concatenated sequences of nad1gene showed thatall strains related to G6 genotype (Figure 4). Moreover, reference sequences for G6 genotype available in the GenBank, were included in the comparative analysis.
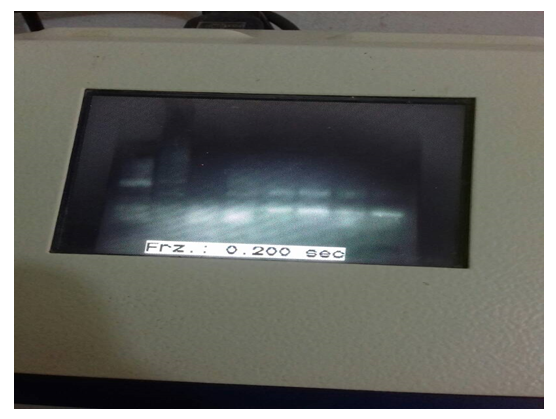
Figure 1: Nested PCR product
Lane 1,2,3,4,5: DNA extracted from echinococcosis cyst sample from patient`s lung (+ve), Lane Nc: negative control, Lane PC: E. granulosus (G6) strain DNA (positive control), MW: molecular weight marker (500bp).
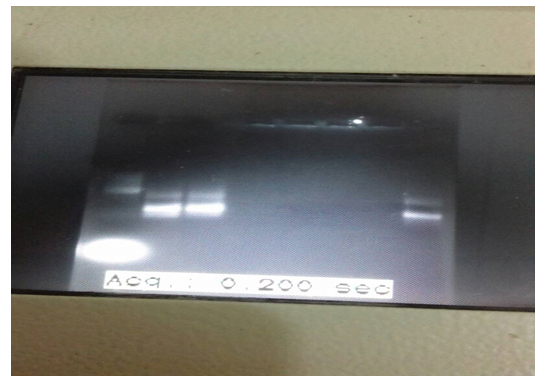
Continued figure (1): nested PCR product for sample 6 and 7 were positive
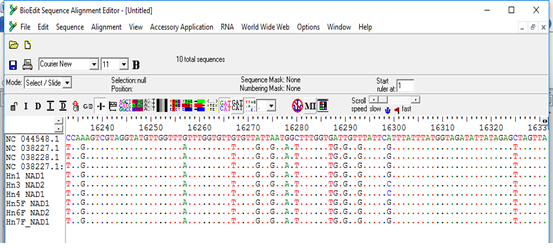
Figure 2: Multiple sequence alignment of nad1 gene of samples and Echinococcus granulosus mitochondrion, complete genome (NC_044548.1), Echinococcus granulosus sensu lato genotype G6 isolate 1 mitochondrion, complete genome (NC_038227.1) and Echinococcus granulosus sensu lato genotype G7 isolate 27 mitochondrion, complete genome (NC_038228.1) as reference samples.
*All samples except 3 and 4 showed nucleotide substitution A>G when compared with complete genome reference sequence, samples 3 and 4 showed nucleotide substitution G>C when compared with G6/7 reference sequence.
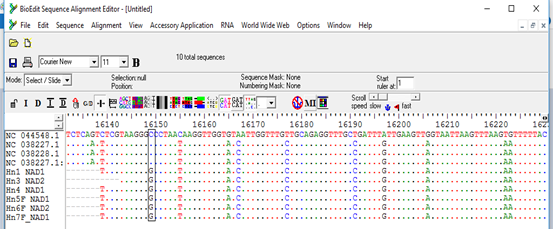
Figure 3: Multiple sequence alignment of nad1 gene of samples and Echinococcus granulosus mitochondrion, complete genome (NC_044548.1), Echinococcus granulosus sensulato genotype G6 isolate 1 mitochondrion, complete genome (NC_038227.1) and Echinococcus granulosus sensulato genotype G7 isolate 27 mitochondrion, complete genome (NC_038228.1) as reference samples.
*All samples showed nucleotide substitution C>G.
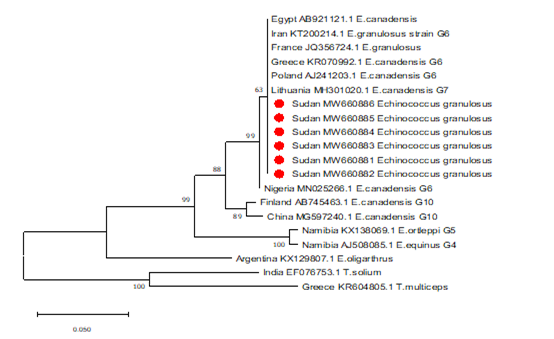
Figure 4: Phylogenetic relationship of Echinococcus granulosus-complex genotypes recovered from Sudanese human and other genotypes identified globally.
Partial NADH dehydrogenase subunit 1 gene sequences generated from EG isolates were aligned with sequences of other EG isolates from different parts of the world. Sequences were analyzed with the BioEdit software (Ibis Biosciences, Carlsbad, CA, USA). The phylogenetic tree was constructed using neighbor joining tree implemented in MEGA software version 7. Bootstrap values were calculated from analysis of 1000 replicates of the data set, and values greater than 50% are indicated at the appropriate nodes. Each EG isolate is designated by country of origin, GenBank accession number and the genotype of the isolate when available. The GenBank accession numbers (MW660881, MW660882, MW660883, MW660884, MW660885 and) were given for the Sudan isolates of genotype 6 (E. canadensis). The strains from Sudan those described in this study are highlighted in red color for clarity. T.solium and T.multiceps were used as the out group sequence data.
4. Discussion
Cystic pulmonary echinococcosis (CE), affects many people throughout the world, although, advances in diagnosis and treatment of CE had been achieved in the recent years. There is still a limit to the disease control. As an endemic region with high incidence of CE, the disease is considered as a public health and socio-economic problem in Sudan. Investigations on the epidemiology and different genotypes of parasites in the intermediate and final hosts should be considered in any endemic area to achieve the evidence based control and management programs [14]. In the African countries, the G6 has been reported as the dominant genotype [15]. The genotype of all isolates from camel in Mauritania, Algeria and Sudan has been reported to be G6 [16,17]. However, other studies in Kenya and Libya have shown a noticeable prevalence of G1 strains in camel isolates [17,18]. Different genotypes of E. granulosus including G1, G 5 and G6 /G7 have been reported from different hosts in Sudan [13,19,20]. The study results showed that all the hydatid cysts analyzed for species/genotypes identification were G6 (camel strain). The exclusive occurrence and a predominant circulation of the camel genotype (G6) suggested that cattle can play an important role in the transmission dynamic and the epidemiology of the disease in Sudan. This study was in agreement with many studies in Sudan conducted to identify the genotypes of E.granulosus in human. Omer et al., Ahmed, Aradaib and Omer et al. documented the predominance of E. canadensis‘camel strain’ (G6) [12,21,22]. However, genotyping of 22 patients isolates from Elshaab Teaching Hospital, Khartoum showed the G6 (Camel strain) in all except one patient showed G1 strain (sheep strain) [23]. As a lot of studies conducted in Sudan for molecular genotyping of E.granulosus that showed the predominant strain among livestock was E. canadensis‘camel strain’ (G6) [13,24,25]. While other studies demonstrated that E. canadensis‘camel strain’ (G6) coexist with E. ortleppi‘cattle strain’ (G5) and E. granulosus senso stricto‘sheep strain’ (G1) are circulating in production animals, where E. ortleppi was found in cattle only in Central, Eastern and western Sudan [12,17,20,26,27]. The exclusive occurrence and a predominant circulation of the camel genotype (G6) suggested that cattle can play an important role in the transmission dynamic and the epidemiology of the disease in Sudan.Unlike the study done in Iran which found all the human cases studied in that study were infected by G1 strain of E. granulosus (sheep strain). This finding is emphasis on the importance of sheep as a source of canine and subsequently human infections which support the idea that G1 strain is predominant in the East Azerbaijan Province [28]. The phylogenetic reconstruction method highly supported the existence of main strains, G6 genotype in humans from Sudan. However , few phylogenetic studies were conducted to determine the phylogenetic relationship of the identified the genotypes of EG in the Sudan. In this study the phylogenetic analysis illustrated that Echinococcus canadensis genotype 6 (G6) is the most infectious and widespread genotypes in Sudan, confirming the results of previous studies [13,20]. Results of our study showed presence of various nucleotide substitution in nad1 genes that are region specific in compare to reference strains in the world. In conclusion, data on frequency of E.granulosus genotypes in human revealed no changing pattern of genotypes distribution between isolates in Sudan. G6 was identified as the most common species in human in Sudan.
References
- Romig T, Ibrahim K, Kern P et al. A molecular survey on cystic Echinococcus in Sinnar area, Blue Nile estate (Sudan). Chin Med J 124 (2011) 2829- 2833.
- Kumsa B. Hydatidosis in Nekemet: prevalence in slaughtered cattle and sheep estimated economic loss and incidence in stray dog. DVM Thesis. Addis Ababa University, Faculty of Veterinary Medicine. Debre Zeit, Ethiopia (1994).
- Moro P and Schantz PM. Echinococcosis: a review. Int J Dis 13(2009): 125-133.
- Thompson RC, McManus DP. Towards a taxonomic revision of the genus Echinococcus. Trends Parasitol 18 (2002): 452-457.
- Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev 17 (2004): 107-135.
- Budke CM, Deplazes P, Torgerson PR. Global socioeconomic impact of cystic echinococcosis. Emerging Infect Dis 12 (2006): 296-303.
- Erneo B, Ochi, David A Akol, et al. A review on epidemiology of hydatidosis in livestock and humans in South Sudan. International Journal of Research Studies in Biosciences 4 (2016): 4-10.
- Abdel-Malik E. Check-list of helminthes parasites of domesticated animals in Sudan. Ind Vet J 36 (1959): 281-286.
- Carmena, Martinez DJ, Benito A, et al. Characterization of excretory-secretory products from protoscoleces of Echinococcus granulosus and evaluation of their potential for immunodiagnosis of human cystic echinococcosis. Parasitology 129 (2004): 371-378.
- Sharafi SM, Rostami-Nejad M, Moazeni M, et al. Echinococcus granulosus genotypes in Iran. GastroenterolHepatol Bed Bench 2 (2014): 82-88.
- Elmahdi IE, Ali QM, Magzoub MMA. Ibrahim, et al. Cystic echinococcosis of livestock and humans incentral Sudan. Ann. Trop. Med. Parasitol 98 (2004): 473-479.
- Omer RA, Dinkel A, Romig T, et al. A molecular survey ofcystic echinococcosis in Sudan. Veterinary Parasitology 169 (2010): 340-346.
- Ibrahim K, Thomas R, Peter K, et al. A molecular survey on cystic echinococcosis in Sinnar area, Blue Nile state (Sudan). Chin Med J 124 (2011): 2829-2833.
- Ebrahimipour M, Sadjjadi SM, Darani HY, et al. Molecular Studies on Cystic Echinococcosis of Camel (Camelus dromedarius) and Report of Echinococcus ortleppi in Iran. IranJ Parasitol 12 (2017): 323-331.
- Sadjjadi SM. Present situation of echinococcosis in the Middle East and arabic north africa. Parasitol Int 55 (2006): 197-202.
- Bardonnet K, Piarroux R, Dia L, et al. Combined ecoepidemiological and molecular biology approaches to assess Echinococcus granulosus transmission to humans in mauritania: Occurrence of the 'camel' strain and human cystic echinococcosis. Trans R Soc Trop Med Hyg 96 (2002): 383-386.
- Dinkel A, Njoroge EM, Zimmermann A, et al. A PCR system for detection ofspecies and genotypes of the Echinococcus granulosus complex, with reference to epidemiological situation in eastern Africa. Int J Parasitol 34 (2004): 645-653.
- Tashani OA, Zhang LH, Boufana B, et al. Epidemiology and straincharacteristics of Echinococcus granulosus in the Benghazi area of eastern Libya. Ann Trop Med Parasitol 96 (2002): 369-81.
- Omer RA, Daugschies A, Gawlowska S, et al. First detection of Echinococcus granulosus sensu stricto (G1) in dogs in central Sudan. Parasitol Res 117 (2018): 1657-1661.
- Mohamed Ahmed, Bashir Salim, Martin P, et al. First molecular characterization of Echinococcus granulosus (sensustricto) genotype 1 among cattle in Sudan. BMC Veterinary Research 14 (2018): 36.
- Omer RA, Dinkel A, Romig T, et al. Strain characterizationof human hydatidosis in Sudan. Int Arch Hydatid 35 (2004): 41.
- Ahmed ME, Aradaib IE. Molecular characterization of Echinococcus granulosus using polymerase chain reaction. Int J Trop Med 1 (2006): 03-32.
- Ahmed ME, Abdelrahim M, Grobusch M, et al. Surgery for Pulmonary Lung Disease in Khartoum,ElShaabTeaching Hospital. Proceeding of the 25th world congress of Echinococcosis in Friendship Hall- Khartoum, Sudan, 23-27 November 2013.
- Osman AM, Ahmed EM, Omer AR, et al. Rapid detection of Echinococcus granulosus - complex and specific identification camel genotype (G6) a nested PCR. Proceeding of the 22nd International Congress of Hydatiology, Athens, Greece (2007): 80-85.
- Elmahdi IE, Elmahdi AE, AbdAlmalaik AAH, et al. Camels Cystic Echinococcosis: Epidemiology and Genotyping of Echinococcus species in Sudan. J Sci and Techno in Agri and Vet Sci 14 (2013): 69-79.
- Elmahdi IE, Elmahdi AE, AbdAlmalaik AA, et al. A molecular survey for genotying of Echinococcus species in Sudan. Proceeding of the 25th world congress of Echinococcosis in friendship hall- Khartoum, Sudan, 23-27 November 2013b.
- Makky A, Ibrahim K, Kern P, et al. Cystic echinococcosis in Camels and cattle in White Nile area, Sudan. Proceeding of the 25th world congress of Echinococcosis in Friendship Hall- Khartoum, Sudan, 23-27 November 2013.
- Vahedi A, Mahdavi M, Ghazanchaei A, et al. Genotypic characteristics of hydatid cysts isolated from humans in East Azerbaijan Province (2011-2013). J Anal Res Clin Med 2 (2014): 152-157.
