Diagnosis of Hairy Cell Leukemia and HCL-like Disorders: Pitfalls to Avoid and the need to Integrate Molecular Data
Article Information
Xavier Troussard*, Pauline Kerneves, Elsa Maitre
Department of Hematology, CHU Caen Normandie, Avenue Côte de Nacre, 14033, Caen Cedex, France
*Corresponding Author: Xavier Troussard, Department of Hematology, CHU Caen Normandie, Avenue Côte de Nacre, 14033, Caen Cedex, France.
Received: 07 June 2022; Accepted: 17 June 2022; Published: 30 June 2022
Citation: Xavier Troussard, Pauline Kerneves, Elsa Maitre. Diagnosis of Hairy Cell Leukemia and HCL-like Disorders: Pitfalls to Avoid and the need to Integrate Molecular Data. Archives of Clinical and Medical Case Reports 6 (2022): 491-500.
Share at FacebookAbstract
The diagnosis of Hairy Cell Leukemia and HCL-like diosrders, including HCL variant (vHCL) and Splenic Diffuse Red Pulp Lymphoma (SDRPL), is based on the examination of the peripheral blood and bone marrow smears allowing the identification of hairy cells and the flow cytometric analysis. The immunologic scoring system, based on the reactivity with each of the four markers (CD11c+, CD25+, CD103 and CD123), usually ranges from 4 (typical of HCL) to 0 (HCL-like disorders). HCL is sometimes confused with other HCL-like disorders. Differentiating all the entities may be challenging due to overlapping features, particularly morphologic and immunophenotypic criteria. Unusual and atypical immunophenotypes are frequently observed: the clinical impact of this variability, if it does exist, remains to demonstrate. In a large cohort of patients, we found a CD103 expression, which was heterogeneous and found only in a fraction of hairy cells in 2/68 cases (3%). We also identified CD5 and CD10 expression in 7/68 (10%) and 12/68 (18%) cases, respectively. Looking for the genetic alterations is crucial: the BRAFV600E mutation was detected in 80-90% of HCL. In cases with BRAF wild type (BRAFWT), the possibility of alternative mutations particularly in exon 11 should be excluded. Conversely to HCL, the BRAFV600E mutation was never identified in HCL-like disorders. Activating mutations in MAP2K1 gene (15q22.1-q22.3) were identified in half of the cases. Recurrent hotspot mutations of U2AF1 encoding a protein belonging to the spliceosome were detected in 15% of vHCL. Twenty-four per-cent of patients with SDRPL presented mutations in CCND3 and/or recurrent mutations or losses in BCOR (gene encoding the BCL6 corepressor). There is no known molecular data in jpHCL. Atypical morphologic and immunophenotypic profiles are frequent and the use of genetic data could be offer a new posssibility for a better classification of HCL and HCL-like disorders.
Keywords
Leukemia; Immunophenotypic; Hairy Cell Leukemia
Article Details
1. Introduction
Classical Hairy Cell Leukemia (HCL) and HCL-like disorders are a very heterogeneous group of mature B-cell Chronic Lympho proliferative Disorders (B-CLPD). HCL is a well-defined entity in the 2017 revision of the World Health Organization (WHO) classification of the tumours of haematopoietic and lymphoid tissues. Conversely, HCL-like disorders including HCL-Variant (vHCL) and Splenic Diffuse Red Pulp Lymphoma (SDRPL) remain provisional entities. The HCL-Japanese variant form (jpHCL) is rare and less well defined: there are common points with vHCL but several aspects are different in terms of morphology of hairy cells, degree of leukocytosis or clinical course. Splenic marginal zone lymphoma with circulating villous lymphoid cells (SMZL) is a distinct and very different pathological entity since the WHO 2008 classification: it is characterized by an expansion of the splenic white pulp with the infiltration of the red pulp. Distinguishing HCL from HCL-like disorders may be difficult due to overlaps. An accurate diagnosis is necessary given that different clinical management is required: the first step in a definitive diagnosis is based on the examination of the peripheral blood and bone marrow smears allowing the identification of hairy cells and a specific Flow Cytometric Analysis (FCA). Molecular studies are usually needed, particularly in complex cases, including cases with transformation and/or progression.
1.1. Cytological and immunophenototypic findings: considerable progress in the diagnosis of HCL and HCL-like disorders
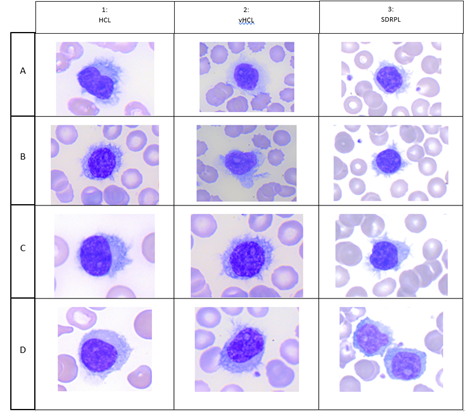
HCL is a rare disease with 1,240 new cases of HCL expected per year in the United States, and 1,417 in the European Union as a whole. Hairy cells are small to medium-sized lymphoid cells with an oval or indented nucleus with homogeneous and spongy chromatin. Nucleoli are typically absent or inconspicuous (Figure 1 A,B,C,D). HCL exhibits a characteristic immunophenotype profile that is strongly positive with pan-B-cell markers (CD19, CD20, CD22) and usually negative for CD5, CD10 and CD23. E Matutes et al proposed a scoring system which considers the reactivity with each of the four markers (CD11c+, CD25+, CD103 and CD123) and gives 1 point if positive and 0 point if negative. Scores range from 4 (typical of HCL) to 0 (HCL-like disorders). 98% of HCL had high score (3 or 4) whereas 88% of HCL-variant and 77% of SLVL scored 1 or 2 and no single case scored 3 or 4 [1]. vHCL represents 800 new incident cases in the United States. The blood picture is monomorphic, with a morphology of the abnormal lymphoid cells intermediate between prolymphocytes and hairy cells. Cells have large and prominent nucleoli with circumferential hair-like cytoplasmic projections (Figure 2, A,B,C,D). CD11c was positive in all cases and CD103 was expressed between two thirds of vHCL cases [2] and 100% positive with 1/35 (3%) patient with partial positivity. Conversely to HCL, CD25 and CD123 are usually negative. In one study, CD123 when present in vHCL (40%) was characteristically dim [3] vHCL may undergo progression and/or transformation occurring in 6% of cases [2]. SDRPL is rare and epidemiologic data are not clearly established. In a cohort of 37 patients with SDRPL, the small to medium-sized abnormal lymphoid cells present a small or not visible nucleolus and some lymphopasmacytoid cells are often observed. Interestingly, CD103 was expressed with different intensities in 13/34 cases (38%), CD11c in 36/37 cases (97%), CD123 in 3/19 cases (16%) and CD25 in 1/37 cases (3%) [4] SDRPL could represent the first step before the occurrence of vHCL: a significant percentage of medium to large cells with a prominent nucleolus was present in 4/37 SDRPL cases with tumoral progression in two cases and transformation in 2 last cases. We present a patient with SDRPL at diagnosis, who progressed to vHCL. As in low grade B-cell lymphomas, the progression of SDRPL in B-cell prolymphocytic leukemia was also reported [5]. jpHCL was identified as a distinct vHCL subtype observed in Japan and possibly could constitute the same disease that SDRPL. In the abnormal lymphoid cells, the nucleolus is usually indistinct. A recent meta-analysis reported 17 jpHCL with circulating lymphoid cells expressing C11c in 12/12 cases, CD103 in 3/12 cases and CD25 in 1/14 cases [6]. SMZL accounts for 2% of lymphoid malignancies: a mixture of heterogeneous cells is identified on the peripheral blood smears with villous cells expressing CD11c but less bright than other splenic lymphomas. CD123 and CD25 are usually negative and CD103 may be dimly positive in rare cases [7].
1.2. Atypical immunologic profiles are frequently observed: pitfalls to avoid
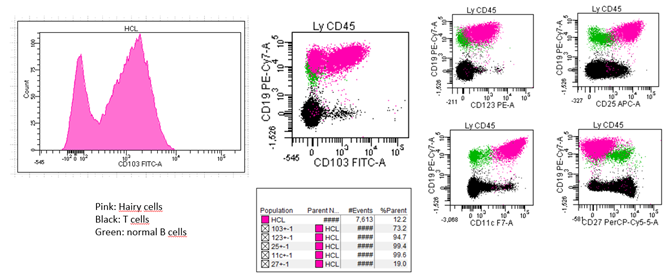
Stefania de Propis et al recently presented the immunophenotypic analysis of 71 consecutive patients with HCL and reported 3 patients with a CD103 expression on only a fraction of hairy cells while expressing CD25, CD11c on all clonal cells and presenting a clonal BRAFV600E mutation and anexin-A1 [8] The 3 patients responded to standard HCL treatment and achieved a Complete Response (CR) after cladribine in 2 cases and interferon-alpha in a patient who refused chemotherapy. All 71 HCL cases were negative for CD5 and CD10. We recently investigated 82 patients including 68 HCL, 5 vHCL/SDRPL and 9 HCL-like disorders Not Otherwise Specified (NOS) because of an uncertain diagnosis after a careful review of peripheral or blood smears and FCA [9]. In this large cohort, an unusual and atypical phenotype was detected in 55 patients. As in the Italian study, we found a CD103 expression which was heterogeneous and found in a fraction of hairy cells in 2/68 cases (3%) (Figure 2). The clinical impact of this variability, if it does exist, remains to demonstrate in a cohort with a higher number of patients. Contrary to the Italian study, we identified CD5 and CD10 expression in 7/68 (10%) and 12/68 (18%) cases, respectively. CD38, CD23 or CD43 varied between bright and dim in 24 (36%), 22 (32%) and 19 (31%) patients tested, respectively. In one another study including 161 patients with HCL, 5 patients (4%) were identified with positive staining for CD5 which generally appears weak and not uniform. CD10 expression was detected in 34/133 cases (26%) [10]. In one another large study analyzing 213 cases including 169 HCL, all HCL cells expressed CD103 and aberrant antigen expression of CD5 and CD10 was present in 2% and 12%, respectively [4]. A patient with CD10 expression without annexin A1 was recently also reported [11]. The differences observed between the different reported studies could be explained by the different procedures of FCA used, the number of events acquired or the type of monoclonal antibody tested. Continued efforts must to be made and It is crucial for the entire medical community to coordinate and standardize the procedures for FCA, as it was done in Chronic Lymphocytic Leukemia (CLL).
1.3. Complex cases requiring extensive Flow Cytometric Analysis (FCA)
In complex cases, accurate diagnosis could be challenging: in that cases, it could be useful to include more informative and validated immunologic markers. Differential mean fluorescence intensity (MFI) of CD43, CD81 and CD200 was proposed to distinguish HCL from vHCL. The median MFI was higher for CD43 and CD200 in HCL and lower for CD81 in vHCL [12]. In our study, CD26 was expressed in 35/36 HCL patients, none of vHCL/SDRPL and only 1/7 HCL-NOS [9]. When adding CD26 to the immunologic HCL scoring system, we demonstrated the specificity was improved suggesting CD26 expression could be informative for distinguishing HCL from HCL-like disorders and better classifying HCL-like disorders [9] Use of a scoring system based on the expression of CD11c, CD22, CD76, CD38 and CD27 was also proposed and appeared to improve the differential diagnosis between SDRPL and SMZL [13].
1.4. Adding molecular studies is required for a definitive diagnosis: BRAFV600E is the molecular hallmark of HCL representing a useful diagnostic possibility and option for therapeutic targeting of BRAF using BRAF Inhibitors (BRAFi)
By Whole-Exome Sequencing (WES) of genomic DNA from purified leukemic cells of one HCL patient, mutation V600E of the BRAF gene (BRAFV600E) as well as CSMD3, SLC5A1, CNTN6 and OR8J1 mutations were identified in 2011 by Enrico Tiacci et al [14]. BRAFV600E was subsequently identified in 48 other HCL patients by Sanger sequencing and was not present in 195 patients with other B-CLPD, suggesting BRAFV600E mutation could be an early and central genetic driver in HCL. The mutation replaces Thymine (T) with adenine (A) in exon 15 of BRAF at position 1799 of the gene coding sequence located in chromosome 7q34. This substitution produces an amino acid change from Valine (V) to glutamate (E) at position 600 (V600E) of the protein sequence and leads to aberrant activation of the protein serine threonine protein kinase B-Raf. The BRAFV600E mutation constitutively activate the mitogen-activated protein kinases-extracellular signal-regulated kinases (MEK–ERK) pathway, leading to enhanced cell proliferation, survival, and ultimately, neoplastic transformation. When using double immunohistochemical or immunofluorescence staining for PAX5 or CD20 and phospho-ERK, phosphorylated ERK was detected in untreated HCL patients with a consistent and dose dependent MEK/ERK dephosphorylation in HCL patients treated with BRAF Inhibitors (BRAFi). In contrast, no effect was observed in any of 10 HCL-like patients, who had much lower basal levels of pMEK/ERK [15]. The authors also demonstrated that in vitro incubation of primary leukemic hairy cells with PLX-4720 (Sorafenib), a BRAFi, led to a marked decrease in phosphorylated MEK and ERK in five additional patients. The BRAFV600E mutation constitutively activates BRAF by autophosphorylation of the protein and downstream MEK–ERK signaling pathway, leading to increased expressions of genes involved in survival and proliferation such as members of the ETS family, FOS, MYC as well of genes involved in MEK/ERK inhibition such as Dual-Specificity Phosphatases (DUSPs). BRAFV600E is detected in 80-90% of HCL. There are definitely HCL cases with BRAF wild type (BRAFWT) [9]. Conversely in HCL-like disorders including vHCL, SDRPL and jpHCL, the BRAFV600E mutation was never identified.
1.5. Consequences of the BRAFV600E mutation
The transcriptional signature which distinguishes HCL from other mature CLPD and close that of memory B-cells is silenced in HCL cells treated by BRAFi. BRAFi treated HCL but not HCL like cells showed a considerable loss of the hairy projections. A consistent loss of viability was also observed when exposing HCL cells in vitro to BRAFi or MEK Inhibitors (MEKi). The in vitro proapoptotic activity of BRAFi was also demonstrated and not identified in none of the 4 HCL-like patients tested [15].
1.6. Alternative mutations
In HCL patients with BRAFWT, the possibility of alternative mutations particularly in exon 11 should be excluded. BRAF mutations were investigated in exon 11 in 24 HCL and 194 various mature B or T-cell neoplasms. Twenty-one patients carried the BRAFV600E mutation in exon 15. Two BRAFWT patients presented BRAF mutations in exon 11 (F468C and D449E). The last patient presented a BRAFV600E mutation associated with a S602T mutation in exon 15 of B-Raf. Two patients showed wild-type sequences for KRAS (exons 2, 3 and 4), NRAS (exons 2, 3 and 4) and HRAS (exons 2 and 3) and in the other patient wild type sequences were found in KRAS (exons 2,3,4), NRAS (exons 2 and 3) and HRAS (exon 2). The functional consequences of the different mutations were not analyzed. All non-HCL lymphomas lacked BRAF mutations [16]. In our experience on 98 patients, including 82 with HCL and 16 with HCL-like disorders, just one patient presented a BRAFF595L mutation without RAS mutations, suggesting the non BRAFV600E mutations are rare in HCL and need to be detected [17]. In absence of BRAFV600E, a single HCL patient was previously reported with a t (7;14) (q34;q32) and an IGH-BRAF fusion associated with the presence of phosphorylated ERK and cyclin D1 by immunohistochemical analyses suggesting the possibility of one another mechanism of activation of the RAS-RAF-MEK-ERK MAP signal transduction cascade [18].
1.7. Other mutations can be detected and could play a role in the progression of the disease
When performing deep sequencing in a large cohort of HCL patients, inactivating KLF2 (19p13.1) mutations were observed in 23% of HCL. KLF2 is a transcription factor controlling the differentiation of multiple B-cell subpopulations, including marginal zone B-cells. In addition to the overexpression of cyclin D1, recurrent inactivation of the cell cycle inhibitor CDKN1B (12p13) was identified 7.5%% of HCL cases. A higher frequency was observed in a series including x patients. TP53 mutations seem infrequent in HCL. Mutations of the genes of the epigenetic regulation were frequently observed, with mutations in the histone methyltransferase KMT2C (MLL3) occurring in 15% of patients and more rarely mutations in histone demethylase KDM6A or histone acetyltransferase CREBBP (CBP). Other mutations in the chromatin remodeling complex family ARID1A, ARID1B were also described [19]. The progression of pre-existing hematologic malignancies of chronic myelomonocytic leukemia, [20, 21] monoclonal B-cell lymphocytosis [22] and Acute Myeloid Leukemia (AML) [23] was reported after the initiation of vemurafenib therapy and was associated with RAS mutations. A progression of CLL was observed in the absence of mutations in RAS [24]. Taken together, these data require a careful monitoring of patients treated by BRAFi.
1.8. BRAFV600E is not specific of HCL: the mutation is usually detected with a lower frequency in solid tumors and other hematologic malignancies
BRAF mutations are found in a wide range of both various solid and hematopoietic tumours with a high incidence of 80% in cutaneous melanoma. Benign naevi of the skin are BRAFV600E, while never experiencing malignant transformation [25]. BRAFV600E is detected in 50% of Langerhans Cell Histiocytosis (LCH) and Erdheim Chester Disease (ECD) and much more rarely in lung, ovarian, bladder, thyroid, prostatic cancers, cholangiocarcinoma or sarcoma/GIST. BRAF mutations have also been identified in other B-CLPD including CLL and multiple myeloma in less than 5% of cases.
1.9. BRAFV600E is missing in HCL cell lines
Seven cell lines including Hair-M, HCLL-7876, EH, Eskol, HC-1 and HCLv-07 have been generated, but evidence linking cell lines to primary tumor remains incomplete. The HCL-associated BRAF mutation was absent in each of the four ANXA1 cell lines (Eskol, Hair-M, HC-1 and HCLL- 7876). The lack of the BRAF mutation in HCLv-07 could be expected because of HCLv origins as well as the absence in the CD5 EH cell line [26]. These data suggest that the model of using cell lines should be limited.
1.10. Cellular origin of HCL
HCL is a fascinating disease and the normal counterpart of hairy cells is still unclear. BRAF gain-of function mutation occurs in earlier differentiation stages including Hematopoietic Stem Cells (HSC) or B-cell lymphoid progenitors of affected HCL patients [27]. When BRAF mutated HSC were transplanted into immunodeficient mice, the mice developed a lethal hematopoietic disorder with splenomegaly and hepatomegaly, anemia and thrombocytopenia, and increases circulating soluble CD25. In contrast, no effect was observed when BRAF mutated B lineage cells were transplanted. A patient with HCL associated with LCH was recently described, suggesting the possibility of a relationship between both diseases. The pattern of distribution of mutant alleles in the mononuclear compartment and bone marrow was clearly different between patients with HCL and LCH/ECD [28]. In LCH/ECD and in the peripheral blood, the majority of mutant alleles were present in CD14+ classical monocytes, CD16+ non-classical monocytes and CD1c positive myeloid dendritic cells. They are also distributed in HSCs and myeloid progenitors in the bone marrow. In HCL, the mutant alleles were not found in monocytes and myeloid cells but were detected in normal B and NK cells. HSCs carrying the BRAF mutation or the mutant (c.172_186del) MAP2K1 deletion transplanted into immunodeficient MISTRG mice induce clusters of medullary, spleen, hepatic, and pulmonary cells whose morphology is that of Langerhans cells (langerine (CD207), CD1a and S100 positive with BRAFV600E mutation and without histological lesion of HCL [29]. Treatment of HCL patients with vemurafenib resulted in normalization of HSPC frequencies and increased myeloid and erythroid output from HSPCs.
1.11. Mutations in HCL-like disorders
When using whole exome sequencing, activating mutations in mitogen-activated protein kinase kinase 1 (MAP2K1) gene (15q22.1-q22.3) were identified in 5/10 samples including 5 vHCL, IGHV4-34 or both. The mutations were associated with TTN mutations in 4 cases, TP53 in 3 cases, ARID1 in 3 cases and U2AF1 in 2 cases [30] In a set of validation of 21 additional samples, Sanger sequencing identified 10 other positive samples leading to the identification of MAP2K1 in 6/15 IGHV4-34 negative vHCL, 4/9 IGHV4-34 positive vHCL and 5/7 IGHV4-34 positive HCL. In the last set of IGHV4-34 negative HCL, just one patient presented MAP2K1 mutation. In that cases, MEKi such as trametinib or binimetinib could be used. TP53 were found in 50% of cases of vHCL. Recurrent hotspot mutations of U2 Small Nuclear RNA Auxiliary Factor 1 (U2AF1) encoding a protein belonging to the spliceosome were detected in 15% of vHCL. Twenty-four per-cent of patients with SDRPL presented mutations in CCND3 and/or recurrent mutations or losses in BCOR (gene encoding the BCL6 corepressor) [31]. In a series of 19 SDRPL patients including 5 with progressive disease, 4 patients presented mutations in NOTCH1 (2 cases), TP53 (1 case) and MAP2KI (1 patient) [32]. KLF2 mutations appear very rare in SDRPL [33]. There is no known molecular data in jpHCL. Integrating molecular studies can be useful in cases of progression of SDRPL to vHCL and for distinguishing HCL from others HCL-like disorders.
2. Conclusion
It could be difficult to distinguish HCL from HCL-like disorders due to overlaps between all the entities. Peripheral blood examination and FCA are very useful tools for the diagnosis of HCL and HCL-like disorders: however atypical profiles are relatively frequent and integration of all the data including morphologic, immune phenotypic and genetic findings is recommended. It will offer new possibilities for a better classification of HCL-like disorders in the future. A collaborative work among scientists and pathologists from different centers is also required and expected in order to harmonize the procedures especially for the diagnosis and also the monitoring of the response to the different treatments used in HCL and HCL-like disorders.
References
- Matutes E, Morilla R, Owusu-Ankomah K, et al. The immunophenotype of Hairy Cell Leukemia (HCL). Proposal for a scoring system to distinguish HCL from B-cell disorders with hairy or villous lymphocytes. Leuk Lymphoma 1 (1994): 57-61.
- Matutes E, Wotherspoon A, Catovsky D. The variant form of hairy-cell leukaemia. Best Pract Res Clin Haematol 16 (2003): 41-56.
- Shao H, Calvo KR, Grönborg M, et al. Distinguishing hairy cell leukemia variant from hairy cell leukemia: development and validation of diagnostic criteria. Leuk Res 37 (2013): 401-409.
- Traverse-Glehen A, Baseggio L, Bauchu EC, et al. Splenic red pulp lymphoma with numerous basophilic villous lymphocytes: a distinct clinicopathologic and molecular entity? Blood 111 (2008): 2253-2260.
- Cheng WY, Zhu YM, Cheng S, et al. Development of B-cell prolymphocytic leukemia in a patient with splenic diffuse red pulp small B-cell lymphoma. Leuk Lymphoma 59 (2018): 1990-1993.
- Ito M, Harada T, Lang L, et al. Hairy Cell Leukemia-Japanese Variant: Report of a Patient and Literature Review. Int J Surg Pathol 2022.
- Donzel M, Baseggio L, Fontaine J, et al. New Insights into the Biology and Diagnosis of Splenic Marginal Zone Lymphomas. Curr Oncol 28 (2021): 3430-3447.
- De Propris MS, Musiu P, Intoppa S, et al. Hairy cell leukaemia with low CD103 expression: A rare but important diagnostic pitfall. Br J Haematol (2022).
- Maitre E, Cornet E, Salaün V, et al. Immunophenotypic Analysis of Hairy Cell Leukemia (HCL) and Hairy Cell Leukemia-like (HCL-like) Disorders. Cancers (Basel) 14 (2022): 1050.
- Robbins BA, Ellison DJ, Spinosa JC, et al. Diagnostic application of two-color flow cytometry in 161 cases of hairy cell leukemia. Blood 82 (1993): 1277-1287.
- Wang HY, Heyman BM. Annexin A1- but CD10+ hairy cell leukemia. Blood 139 (2022): 1924.
- Salem DA, Scott D, McCoy CS, et al. Differential Expression of CD43, CD81, and CD200 in Classic Versus Variant Hairy Cell Leukemia. Cytometry B Clin Cytom 96 (2019): 275-282.
- Baseggio L, Traverse-Glehen A, Callet-Bauchu E, et al. Relevance of a scoring system including CD11c expression in the identification of splenic diffuse red pulp small B-cell lymphoma (SRPL). Hematol Oncol 29 (2011): 47-51.
- Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med 364 (2011): 2305-15.
- Pettirossi V, Santi A, Imperi E, et al. BRAF inhibitors reverse the unique molecular signature and phenotype of hairy cell leukemia and exert potent antileukemic activity. Blood 125 (2015): 1207-1216.
- Tschernitz S, Flossbach L, Bonengel M, et al. Alternative BRAF mutations in BRAF V600E-negative hairy cell leukaemias. Br J Haematol 165 (2014): 529-533.
- Maitre E, Tomowiak C, Lebecque B, et al. Deciphering Genetic Alterations of Hairy Cell Leukemia and Hairy Cell Leukemia-like Disorders in 98 Patients. Cancers 14 (2022): 1904.
- Thompson ER, Lim KJC, Kuzich JA, et al. Detection of an IGH-BRAF fusion in a patient with BRAF Val600Glu negative hairy cell leukemia. Leuk Lymphoma 61 (2020): 2024-2026.
- Oscier D, Stamatopoulos K, Mirandari A, et al. The Genomics of Hairy Cell Leukaemia and Splenic Diffuse Red Pulp Lymphoma. Cancers (Basel) 14 (2022): 697.
- Callahan MK, Rampal R, Harding JJ, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med 367 (2012): 2316-2321.
- Abdel-Wahab O, Klimek VM, Gaskell AA, et al. Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF- and NRAS-mutant malignancies. Cancer Discov 4 (2014): 538-545.
- Simnica D, Ittrich H, Bockemeyer C, et al. Targeting the Mutational Landscape of Bystander Cells: Drug-Promoted Blood Cancer From High-Prevalence Pre-neoplasias in Patients on BRAF Inhibitors. Front Oncol 10 (2020): 540030.
- Dietrich S, Pircher A, Endris V, et al. BRAF inhibition in hairy cell leukemia with low-dose vemurafenib. Blood 127 (2016): 2847-2855.
- Yaktapour N, Meiss F, Mastroianni J, et al. BRAF inhibitor-associated ERK activation drives development of chronic lymphocytic leukemia. J Clin Invest 124 (2014): 5074-5084.
- Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet 33 (2003): 19-20.
- Tiacci E, Pucciarini A, Bigerna B, et al. Absence of BRAF-V600E in the human cell lines BONNA-12, ESKOL, HAIR-M, and HC-1 questions their origin from hairy cell leukemia. Blood 119 (2012): 5332-5333.
- Chung SS, Kim E, Park JH, et al. Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med 6 (2014): 238ra71.
- Milne P, Bigley V, Bacon CM, et al. Hematopoietic origin of Langerhans cell histiocytosis and Erdheim-Chester disease in adults. Blood 130 (2017): 167-175.
- Rafiei A, Wilk CM, Helbling PM, et al. BRAFV 600E or mutant MAP2K1 human CD34+ cells establish Langerhans cell-like histiocytosis in immune-deficient mice. Blood Adv 4 (2020): 4912-4917.
- Waterfall JJ, Arons E, Walker RL, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet 46 (2014): 8-10.
- Curiel-Olmo S, Mondéjar R, Almaraz C, et al. Splenic diffuse red pulp small B-cell lymphoma displays increased expression of cyclin D3 and recurrent CCND3 mutations. Blood 129 (2017): 1042-1045.
- Martinez D, Navarro A, Martinez-Trillos A, et al. NOTCH1, TP53, and MAP2K1 Mutations in Splenic Diffuse Red Pulp Small B-cell Lymphoma Are Associated With Progressive Disease. Am J Surg Pathol 40 (2016): 192-201.
- Jallades L, Baseggio L, Sujobert P, et al. Exome sequencing identifies recurrent BCOR alterations and the absence of KLF2, TNFAIP3 and MYD88 mutations in splenic diffuse red pulp small B-cell lymphoma. Haematologica 102 (2017): 1758-1766.