Crucial Biomarkers for Pulmonary Arterial Hypertension (PAH) by Transcriptome Comparison with Idiopathic Pulmonary Fibrosis with and without PH and Identification of Essential Signaling Pathways- A Meta Analysis and Bioinformatics Study
Article Information
Bhuvnesh Rai*
Department of Molecular Medicine and Biotechnology, Sanjay Gandhi Postgraduate Institute of Medical Sciences,Lucknow, 226014, India
*Corresponding Author: Bhuvnesh Rai, Department of molecular medicine and biotechnology, Sanjay Gandhi Postgradutae Institute of Medical Sciences, Lucknow-226014, India
Received: 14 June 2021; Accepted: 19 July 2021; Published: 21 July 2021
Citation: Bhuvnesh Rai. Crucial Biomarkers for Pulmonary Arterial Hypertension (PAH) by Transcriptome Comparison with Idiopathic Pulmonary Fibrosis with and without PH and Identification of Essential Signaling Pathways- A Meta Analysis and Bioinformatics Study. Journal of Bioinformatics and Systems Biology 4 (2021): 74-102.
Share at FacebookAbstract
The development of pulmonary arterial hypertension (group I PH) complicates many interstitial lung diseases, including idiopathic pulmonary fibrosis (IPF) mostly present with underlined pulmonary hypertension, is suspected to be an independent risk factor for mortality in chronic lung diseases. This meta-analysis of transcriptomics study of pulmonary arterial hypertension and pulmonary fibrosis associated with and without pulmonary hypertension aims to utilize current evidences to extract novel genetic identifiers specifically for PAH in order to identify it among all 5 groups of pulmonary hypertension in IPF patients using pre existing vast number of observational experimental databases freely accessible publicly to facilitate early diagnosis of PAH and thereby improving its therapeutics. This meta-analysis framework extracts expression intensity features from each study, corresponding to genes that are consistently among the highly significant differentially expressed genes (DEGs) in PAH and IPF (with and without PH).
Keywords
Pulmonary arterial hypertension; Idiopathic pulmonary fibrosis; Microarray; DEGs; Biomarker; Transcriptomics; Databases
Pulmonary arterial hypertension articles; Idiopathic pulmonary fibrosis articles; Microarray articles; DEGs articles; Biomarker articles; Transcriptomics articles; Databases articles
Pulmonary arterial hypertension articles Pulmonary arterial hypertension Research articles Pulmonary arterial hypertension review articles Pulmonary arterial hypertension PubMed articles Pulmonary arterial hypertension PubMed Central articles Pulmonary arterial hypertension 2023 articles Pulmonary arterial hypertension 2024 articles Pulmonary arterial hypertension Scopus articles Pulmonary arterial hypertension impact factor journals Pulmonary arterial hypertension Scopus journals Pulmonary arterial hypertension PubMed journals Pulmonary arterial hypertension medical journals Pulmonary arterial hypertension free journals Pulmonary arterial hypertension best journals Pulmonary arterial hypertension top journals Pulmonary arterial hypertension free medical journals Pulmonary arterial hypertension famous journals Pulmonary arterial hypertension Google Scholar indexed journals Idiopathic pulmonary fibrosis articles Idiopathic pulmonary fibrosis Research articles Idiopathic pulmonary fibrosis review articles Idiopathic pulmonary fibrosis PubMed articles Idiopathic pulmonary fibrosis PubMed Central articles Idiopathic pulmonary fibrosis 2023 articles Idiopathic pulmonary fibrosis 2024 articles Idiopathic pulmonary fibrosis Scopus articles Idiopathic pulmonary fibrosis impact factor journals Idiopathic pulmonary fibrosis Scopus journals Idiopathic pulmonary fibrosis PubMed journals Idiopathic pulmonary fibrosis medical journals Idiopathic pulmonary fibrosis free journals Idiopathic pulmonary fibrosis best journals Idiopathic pulmonary fibrosis top journals Idiopathic pulmonary fibrosis free medical journals Idiopathic pulmonary fibrosis famous journals Idiopathic pulmonary fibrosis Google Scholar indexed journals Microarray articles Microarray Research articles Microarray review articles Microarray PubMed articles Microarray PubMed Central articles Microarray 2023 articles Microarray 2024 articles Microarray Scopus articles Microarray impact factor journals Microarray Scopus journals Microarray PubMed journals Microarray medical journals Microarray free journals Microarray best journals Microarray top journals Microarray free medical journals Microarray famous journals Microarray Google Scholar indexed journals DEGs articles DEGs Research articles DEGs review articles DEGs PubMed articles DEGs PubMed Central articles DEGs 2023 articles DEGs 2024 articles DEGs Scopus articles DEGs impact factor journals DEGs Scopus journals DEGs PubMed journals DEGs medical journals DEGs free journals DEGs best journals DEGs top journals DEGs free medical journals DEGs famous journals DEGs Google Scholar indexed journals Biomarker articles Biomarker Research articles Biomarker review articles Biomarker PubMed articles Biomarker PubMed Central articles Biomarker 2023 articles Biomarker 2024 articles Biomarker Scopus articles Biomarker impact factor journals Biomarker Scopus journals Biomarker PubMed journals Biomarker medical journals Biomarker free journals Biomarker best journals Biomarker top journals Biomarker free medical journals Biomarker famous journals Biomarker Google Scholar indexed journals Transcriptomics articles Transcriptomics Research articles Transcriptomics review articles Transcriptomics PubMed articles Transcriptomics PubMed Central articles Transcriptomics 2023 articles Transcriptomics 2024 articles Transcriptomics Scopus articles Transcriptomics impact factor journals Transcriptomics Scopus journals Transcriptomics PubMed journals Transcriptomics medical journals Transcriptomics free journals Transcriptomics best journals Transcriptomics top journals Transcriptomics free medical journals Transcriptomics famous journals Transcriptomics Google Scholar indexed journals Databases articles Databases Research articles Databases review articles Databases PubMed articles Databases PubMed Central articles Databases 2023 articles Databases 2024 articles Databases Scopus articles Databases impact factor journals Databases Scopus journals Databases PubMed journals Databases medical journals Databases free journals Databases best journals Databases top journals Databases free medical journals Databases famous journals Databases Google Scholar indexed journals PAH articles PAH Research articles PAH review articles PAH PubMed articles PAH PubMed Central articles PAH 2023 articles PAH 2024 articles PAH Scopus articles PAH impact factor journals PAH Scopus journals PAH PubMed journals PAH medical journals PAH free journals PAH best journals PAH top journals PAH free medical journals PAH famous journals PAH Google Scholar indexed journals idiopathic pulmonary fibrosis articles idiopathic pulmonary fibrosis Research articles idiopathic pulmonary fibrosis review articles idiopathic pulmonary fibrosis PubMed articles idiopathic pulmonary fibrosis PubMed Central articles idiopathic pulmonary fibrosis 2023 articles idiopathic pulmonary fibrosis 2024 articles idiopathic pulmonary fibrosis Scopus articles idiopathic pulmonary fibrosis impact factor journals idiopathic pulmonary fibrosis Scopus journals idiopathic pulmonary fibrosis PubMed journals idiopathic pulmonary fibrosis medical journals idiopathic pulmonary fibrosis free journals idiopathic pulmonary fibrosis best journals idiopathic pulmonary fibrosis top journals idiopathic pulmonary fibrosis free medical journals idiopathic pulmonary fibrosis famous journals idiopathic pulmonary fibrosis Google Scholar indexed journals
Article Details
1. Introduction
Many types of diffuse parenchymal lung diseases may cause pulmonary arterial hypertension (PAH) to develop. Arcasoy and associates identified PAH in one quarter of patients who were referred for transplantation with various kinds of advanced lung diseases [1]. Pulmonary arterial hypertension is characterised as a persistent elevation of pulmonary arterial pressure at rest to more than 25 mm Hg or with exercise to more than 30 mm Hg, with a mean pulmonary-capillary wedge pressure and a left ventricular end-diastolic pressure of less than 15 mm Hg, being used as the diagnostic criteria as in the National Institutes of Health (NIH) registry [2]. Pulmonary Arterial Hypertension (PAH) refers to Category I PH containing idiopathic or heritable sources in which the lung vasculature is compromised but not the lung parenchyma. Among the environmental factors associated with increased risk of pulmonary arterial hypertension production, three-hypoxia, anorexigens, and stimulants to the central nervous system-have possible mechanistic underpinnings. PAH has been correlated with some of the coexisting conditions too. Those with possible mechanical references include scleroderma, HIV infection, human herpesvirus (HHV), portal hypertension, thrombocytosis, hemoglobinopathy, and hereditary hemorrhagic telangiectasis. In all of these cases the histological presentation of lung tissue is similar: intimate fibrosis, increased medial thickness, pulmonary arteriolar occlusion and plexiform lesions predominate. Vasoconstriction, smooth muscle cell and endothelial cell proliferation, and thrombosis are the principal vascular modifications in pulmonary arterial hypertension. PAH has been identified as occurring in 5 to 38 percent of scleroderma patients, 4.3 to 43 percent of systemic lupus erythematosus patients, and 21 percent of rheumatoid arthritis patients [3, 4]. Sarcoidosis is also associated with PAH in 1 to 28 percent of cases, which is more common in more advanced disease patients [5].
PAH in patients with idiopathic pulmonary fibrosis (IPF) has been documented but the prevalence has not been well-defined. In an Unified analyses Organ exchange registry network, Shorr and colleagues found that about one-quarter of 2,000 IPF patients diagnosed for lung transplants, had PAH [6, 7]. Idiopathic pulmonary fibrosis (IPF), also known as cryptogenic fibrosing alveolitis, is a clinicopathologic term referring to an unexplained cause typically fatal condition characterised by varying degrees of inflammation and fibrosis in the parenchyma of the lungs [8]. Mean survival in IPF has been estimated to be 3 to 6 yr but with a variable clinical course. IPF patients' pathological analysis of lung specimens display a variety of histological patterns. Normal interstitial pneumonia (UIP) is a particular histological pattern of interstitial fibrosing pneumonia seen in most IPF-patients. UIP is the hallmark trait of IPF histopathology, characteristics include temporal and spatially heterogeneous fibrosis, clusters of fibroblasts and myofibroblasts (fibroblastic foci), and excessive deposition of disorganized collagen and extracellular matrix (ECM), resulting in distortion of normal lung morphology, with or without a cyst formation [9].
An important complication of chronic lung diseases here specifically IPF, that is strongly linked to the mortality, is the presence of pulmonary hypertension (PH). The prevalence of PH among patients with IPF depends on the IPF severity. PH affects < 10 per cent of patients with IPF in the early stages or when first diagnosed. However, the frequency of PH rises markedly as the IPF progresses. An occurrence of 32% was identified in one study of patients undergoing lung transplantation and thus in an advanced stage of IPF. Subsequent studies have provided significant support to percentage increase to between 32 and 50 percent [10-12]. It is important to note, though, that the signs for PH and IPF are very close (shortness of breath and exertional dyspnea) and, as such, under-diagnosis of PH in patients with IPF is practicable. PH is characterised by a mean pulmonary arterial pressure (mPAP) of approximately >25mmHg and a pulmonary artery coil pressure (PAWP) of <15mmHg and elevated pulmonary vascular resistance (PVR) > 3 wood units (WU) [10]. The pathological process in PH is characterised by extensive vascular remodelling including increased proliferation of smooth muscle cells (PASMC) in the pulmonary artery. [13, 14] This results in vessel lumen narrowing and obliterating resulting in improved vascular tone. About the same way, regardless of the elevated pressure of the pulmonary vasculature, the right ventricle (RV) helps to brace for remodelling, hypertrophy, inflammation and finally right-sided cardiac failure and death [15]. PH is subdivided into 5 comprehensive subsets of PH: Group I – Group V PH. Group I PH comprises idiopathic or heritable pulmonary arterial hypertension (PAH) where the lung vasculature is affected but not the lung parenchyma. Class II PH is related to left heart disease. Group III PH is associated with chronic lung diseases which affect parenchyma and hypoxemia in the lungs. Group IV PH is chronic pulmonary thromboembolic hypertension (CTEPH); Group V PH finally includes PH from unclear and multifactorial mechanisms.
We hypothesized that PAH is common in patients with more advanced IPF and may be an independent risk factor for mortality. We attempted to define this association using a cohort of patients respective for IPF and PAH who underwent lung biopsies as part of their evaluation using their biopsy tissue as the study sample. We propose a comprehensive meta-analysis for the overall study effect size Z score , P value (P<0.05) and hetrogenity (I2 <50%) method that establishes global relations between transcriptomics studies without publication bias by the use of review manager tool (Revman 5.4) and further, analysis of defined groups by the transcriptome analysis console (TAC 4.0). Our architecture uses this method to extract gene from any global data set function in research that is correlated with genes most commonly or differentially expressed in studies of PAH and PF (with and without PH) providing new insights into novel PAH genetic biomarkers and thereby improving its future therapeutics.
2. Methods and Materials
2.1 Data collection
Data-sets were searched pertaining to pulmonary arterial hypertension (PAH) and pulmonary fibrosis associated gene expression on gene expression omnibus (GEO, NCBI) databases. Two PAH data sets (gse113439, gse53408) [16, 17, 18] with lung biopsy tissue as the sample source were identified focusing primarily on PAH gene profiling. Additionally, two PF data sets (gse24988, gse19976) [19, 20] also with lungs biopsy samples were obtained, for cross reference and comparison. All the datasets shared a common platform (homosapiens & Affymetrix Human Gene 1.0 ST Array). The detailed information about the selected data sets is provided in (Table 1). In total, we
curated gene expression data from these four publicly available data sets keeping the study independent of age, gender, race and region.
|
S. no. |
Study accession no. |
Disease Acronym |
Study description |
No. of samples (disease specific) |
Species and platform (GPL6244) |
|
2 |
GSE113439 |
PAH |
Gene expression profiling of pulmonary arterial hypertension |
15 |
Affymetrix Human Gene 1.0 ST Array |
|
3 |
GSE24988 |
1-PF with PH 2-PF with no PH |
Gene expression profiles based on Pulmonary Artery Pressures in Pulmonary Fibrosis |
62 30 |
Affymetrix Human Gene 1.0 ST Array |
|
4 |
GSE19976 |
PF |
Gene expression analysis of lung biopsies from patients with two different forms of pulmonary sarcoidosis |
8 |
Affymetrix Human Gene 1.0 ST Array |
|
5 |
GSE53408 |
PAH |
Metabolomic heterogeneity of severe pulmonary arterial hypertension |
12 |
Affymetrix Human Gene 1.0 ST Array |
|
6 |
GSE113439, GSE24988, GSE19976, GSE53408 |
PAH vs.PF with PH and PF with no PH base line contr ol (healthy lung biopsy tissue ) |
Meta analysis with transcriptome study TAC 4.0 significant analysis microarray |
PAH-22 PF with PH-22 PF with no PH-22 Healthy control-22 Total=88 |
Affymetrix Human Gene 1.0 ST Array |
Table 1: Human datasets included for transcriptomic analysis.
2.2 Meta-analysis work flow
An equivalent protocol was used to evaluate all of the findings. Until doing the TAC analysis for the differentially expressed genes and the REACTOME pathway analysis, each study was pre-processed including quality management and standardization by REVMAN 5.4. Using uniform threshold condition false discovery rate (FDR) f-test < 1E-43, differentially expressed genes were incorporated by TAC 4.0 similarly, associated pathways were identified using threshold P value (P<0.05) and gene ontology of DEGs. in PAH was identified by PANTHER (Figure 1).
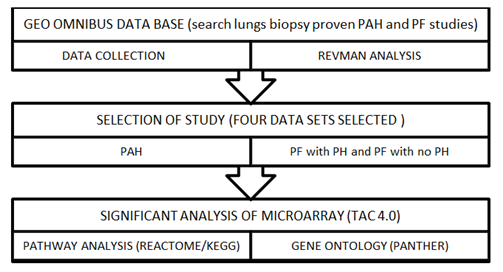
Figure 1: Work flow of meta-analysis process.
2.3 Statistical meta-analysis: Rev-man manager 5.4
The software RevMan 5.4 was used for statistical analysis of all datasets. Dichotomous data was analyzed using the statistical method Mental Haenszel and the model of fixed effect analysis. The data was measured with 95 percent analysis complete as well as total CI (Confidence interval) with effect measures OR (odds ratio). The heterogeneity was measured by (I2 < 50%) with P value (P< 0.05) and overall effect size was measured by Z score with P value (P<0.05). Represented by Funnel and Forest plots.
2.4 Microarray analysis for identification of DEGs in PAH vs. IPF with and without PH
The analysis was performed by TAC 4.0 software along with background adjustment, quantile normalization, summarization, and log2 value transformation using RMA+DABG algorithm. At first principal component analysis (PCA) was executed to obtain the overall similarities and dissimilarities of the log-transformed expression ratios of genes between all the samples of all the three groups. Further, ANOVA was used for the statistical evaluation among groups (PAH, PF with PH, PF without PH and control for normalization). Subsequently all statistically evaluated genes were sorted to obtain the significant ones with a cut off condition FDR F Test (f<1E-43) for the differential gene expression study and generation of hierarchical clustering using distance metric (Euclidean distance). Distances between clusters of objects were computed using the complete linkage method. Sample used in each category were (PAH-22, PF with PH -22, PF without PH-22 and CONTROL-22).
2.5 Quality check through Box plot by TAC
Boxplot displays the distribution of data based on five parameters i.e. (minimum, first quartile (Q1), median, third quartile (Q3), and maximum). It tells about outliers and its values. It also tells symmetry of data, grouping of data, and skewing of data.
2.6 Pathway analysis of DEGs using REACTOME
The open source program Reactome [15, 21] was used to classify particular pathway correlated with separate and typical DEGs in benjamini and hochberg FDR (P < 0,05) classes in PAH and PF.
2.7 Gene Ontology of DEGs using PANTHER
In order to predict gene ontology for novel genes found in PAH and PF groups, PANTHER(Protein ANalysis THrough Evolutionary Relationships) was used, the systematic method integrating genomes, gene function classifications, pathways and methods of statistical analysis, was used to analyze the large-scale genome wide experimental results.
3. Results
3.1 PAH and IPF (with and without PH) microarray datasets
We identified, and included, 4 datasets from PAH and IPF that matched our criteria. Of the 4 datasets, two were from PAH sets (gse113439, gse53408) and 2 were from PF (with and without PH) (gse24988, gse19976) as detailed in table 1.
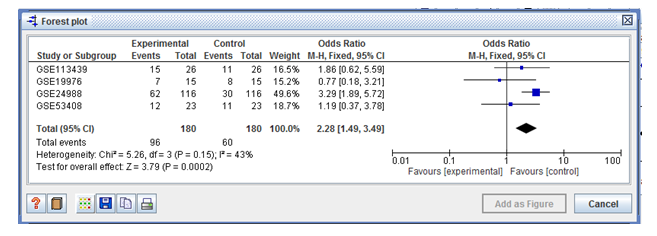
Figure 2: Forest plot shows test for overall effect size z score (Z=3.79) with P value (P=0.0002) and hetrogenity (I2=43%).
3.2 Statistical analysis showing significance of all datasets by REVMAN 5.4
In the forest plots shown with two columns, the left column lists the names in chronological order of the studies wereas, the right column is a chart of the effect calculation (i.e. probability ratio) for each of the experiments, and the horizontal lines represent the intervals of confidence. The graph was plotted on a regular logarithmic scale using odds ratios, so that the confidence intervals are symmetrical around the mean of each sample, and excessive
importance is not given to odds ratios greater than 1 over less than 1. In meta-analysis, the area of each square represents the relative weight of the individual sample. The total meta-analyzed impact measure was depicted as a diamond on the diagram, the lateral points of which suggested intervals of confidence for this calculation. Our plot shows test for overall effect Z score (Z=3.79), P value (P=0.0002) and heterogeneity (I2=43%) (Figure2).
A scatter plot of the impact estimation size from individual experiments is seen in the Funnel plot. The standard error of the impact calculation was used as the indicator of sample size and was plotted on the vertical axis with a reversed scale that put the bigger, more effective studies at the top. The impact results from smaller experiments scattered more uniformly at the right. The outer dashed lines indicated the triangular region within which 95% of studies are expected to lie in the absence of both biases and heterogeneity (fixed effect summary log odds ratio ±1.96× standard error of summary log odds ratio) (Figure3).
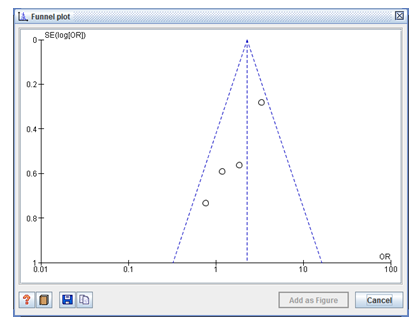
Figure 3: The outer dashed lines in the symmetrical funnel plot indicate the triangular region within which 95% of studies are expected to lie in the absence of both biases and heterogeneity. Funnel plot evaluates the standard error plotted on the vertical axis with a reversed scale showing the larger, most powerful studies towards the top and smaller studies scattered widely at the bottom.
3.3 Quality check of raw data (.CEL) files in PAH and PF (with PH and PF without PH)
Box plot of 88 samples showed the distribution of data to be homogenous in each group (PAH, PF with PH, PF without PH and control) with values significantly lying between (7.2-8) for all the groups. No outliers are present in our study (Figure 4).
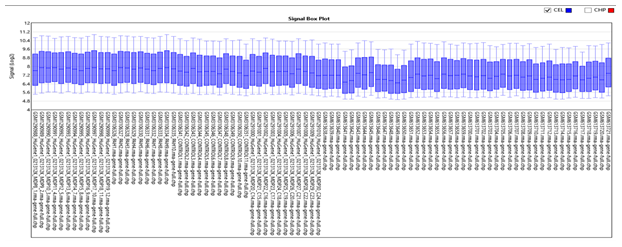
Figure 4: Box plot of 88 samples in PAH vs. PF. It shows that the input sample in each chip array is homogeneous significantly lying between (7.2-8.0).
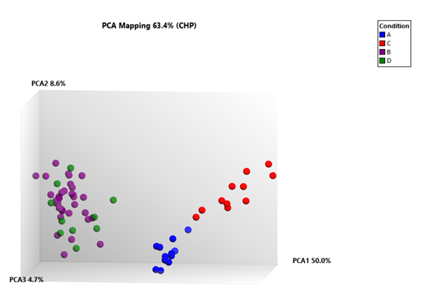
Figure 5A: Three-dimensional principal component analysis (PCA) plot of gene set mapping shows distinction between the 88 samples,.Total of 63.4% variance between PAH (n=22), PF with PH (n=22), PF with no PH and healthy lung tissue samples (n=22) is shown using component 1 (PCA1, 50.0%), component 2 (PCA2, 8.6%), and component 3 (PCA3, 4.7%).The three axes represent the first three principal components identified by the analysis, Each red (C) spot represents a control sample, PAH tissue sample (A), blue spots, and purple spots sample from PF with PH tissue (B) and PF with no PH sample (D), green spots.
3.4 PCA and Heatmap
Principal component analysis plot of gene set mapping showed distinction between the total 88 samples. Total of 63.4% variance between PAH (n=22), PF (with PH) (n=22), PF (without PH) (n=22), and healthy control (n=22) was shown by component 1 (PCA1, 50.0%), component 2 (PCA2, 8.6 %), and component 3 (PCA3, 4.7%). (Figure 5A)
Hierarchical cluster analysis of the significant 353 DEGs filtered out by applying false discovery rate (FDR) f-test < 1E-43 was obtained excluding unassigned genes with log2 fold expression and transcripts. (Figure 5B )
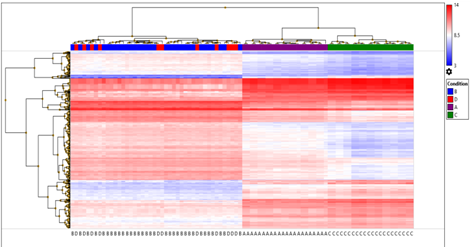
Figure 5B: Heat map and dendogram shows, hierarchical cluster analysis of the significant 353 DEGs. filtered out on giving FDR condition F test (F < 1E-43) excluding unassigned genes with log2 fold expression and transcripts in four sets of samples; A, B, C and D. The clustering was performed through (TAC) 4.0, Distance metric used between objects was the Euclidean distance. Distances between clusters of objects were computed using the complete linkage method.
3.5 Total number of DEGs differentially regulated in PAH and PF (with PH and PF without PH)
For an outline of expression profiles in lung disorder with PAH in human subjects, we used TAC4.0 software to spot differentially expressed genes (DEGs). For the analysis samples from two PAH datasets were merged giving 27 samples in total. Similarly on merging the 2 PF datasets a total of 62 samples were obtained belonging to PF with PH and 22 samples of PF without PH. Merging control samples from all 4 datasets gave a total of 29 samples. Finally 22 samples from each group (PAH, PF with PH, PF without PH and Control for normalization) were analyzed for DEGs.
On comparing PAH vs. PF with PH and PF without PH group we obtained a total of 353 DEGs on applying significant filters i.e. condition F Test (f <1E-43) out of which all 353 DEGs were assigned with gene symbol and included in the study (Table 2).
|
S.no. |
Study accession no. |
Disease acronym |
Significantly gene upregulated Condition fdr f<1E-43 |
Significantly gene downregulated Condition fdr f<1E-43 |
|
1 |
GSE113439+ GSE24988+ GSE19976+ GSE53408 |
PAH vs.PF with PH and PF with no PH base line control (healthy lung biopsy tissue ) Commonly regulated |
198 |
145 |
|
2 |
GSE113439+ GSE24988+ GSE19976+ GSE53408 |
PAH vs.PF with PH and PF with no PH base line control (healthy lung biopsy tissue ) differentially regulated |
5 |
5 |
Table 2: Shows significantly differentially expressed gene which are commonly and differentially up and downregulated in PAH vs. PF with PH and without PH.
From a total of 353 our analysis indicated 145 DEGs commonly downregulated in PAH vs. PF (with PH and PF without PH) (Table 3i) and 198 DEGs were commonly upregulated (Table 3ii ). However, on comparing the three groups 10 DEGs. showed differential regulation significantly in PAH (Table 4).
|
S.no. |
Gene Symbol |
A vs C Fold Change |
B vs C Fold Change |
D vs C Fold Change |
Condition FDR F-Test |
|
1 |
AKT1 |
-1.28 |
-2.94 |
-2.85 |
8.83E-44 |
|
2 |
ARF5; |
-1.96 |
-3.17 |
-3.29 |
1.40E-45 |
|
3 |
ATP5G1 |
-1.34 |
-9.28 |
-8.1 |
8.83E-44 |
|
4 |
ATRAID |
-1.71 |
-3.85 |
-3.86 |
2.80E-45 |
|
5 |
BRK1 |
-1.38 |
-3.93 |
-3.9 |
1.40E-45 |
|
6 |
BRK1 |
-1.17 |
-3.42 |
-3.65 |
1.40E-45 |
|
7 |
C12orf10 |
-1.56 |
-3.05 |
-3.03 |
1.40E-45 |
|
8 |
C20orf24 |
-1.09 |
-3.79 |
-3.42 |
1.40E-45 |
|
9 |
CCDC130 |
-1.45 |
-2.59 |
-2.42 |
1.40E-45 |
|
10 |
CCDC97 |
-1.25 |
-2.42 |
-2.31 |
1.40E-45 |
|
11 |
CCND3 |
-1.48 |
-4.02 |
-3.91 |
1.40E-45 |
|
12 |
CDC34 |
-1.65 |
-3.89 |
-3.56 |
1.40E-45 |
|
13 |
CEBPD |
-1.41 |
-4.88 |
-4.74 |
1.40E-45 |
|
14 |
COX5B |
-1.82 |
-8.54 |
-7.87 |
1.40E-45 |
|
15 |
CRTC3 |
-1.28 |
-2.72 |
-2.51 |
1.40E-45 |
|
16 |
DIRC2 |
-1.38 |
-3.05 |
-3.01 |
1.40E-45 |
|
17 |
ERH |
-1.15 |
-3.05 |
-3.24 |
1.40E-45 |
|
18 |
FKBP1C |
-1.28 |
-3.04 |
-2.88 |
1.40E-45 |
|
19 |
GABARAP |
-1.39 |
-3.09 |
-3.24 |
8.41E-45 |
|
20 |
GABARAPL1 |
-1.18 |
-3.93 |
-3.98 |
1.40E-45 |
|
21 |
GDI1 |
-1.46 |
-2.47 |
-2.4 |
1.40E-45 |
|
22 |
GIT1 |
-1.35 |
-2.47 |
-2.54 |
2.80E-45 |
|
23 |
GMPR |
-1.07 |
-4.06 |
-4.2 |
1.40E-45 |
|
24 |
GNA11 |
-1.48 |
-3.36 |
-3.17 |
1.40E-45 |
|
25 |
GNAI2 |
-1.17 |
-1.95 |
-1.83 |
1.40E-45 |
|
26 |
GNB2 |
-1.29 |
-2.4 |
-2.33 |
1.40E-45 |
|
27 |
GSK3A |
-1.14 |
-2.34 |
-2.34 |
1.40E-44 |
|
28 |
HEBP1 |
-1.11 |
-3.25 |
-3.28 |
1.40E-45 |
|
29 |
HINT2 |
-1.72 |
-5.98 |
-5.8 |
1.96E-44 |
|
30 |
HIST1H1E |
-1.28 |
-4.63 |
-4.96 |
1.96E-44 |
|
31 |
HIST2H2BA |
-1.45 |
-2.97 |
-3.02 |
2.10E-44 |
|
32 |
HNRNPUL1 |
-1.19 |
-2 |
-2 |
1.40E-45 |
|
33 |
ICAM2 |
-2.05 |
-6.76 |
-5.88 |
1.40E-45 |
|
34 |
IDH3G |
-1.27 |
-3.05 |
-2.88 |
4.20E-45 |
|
35 |
INF2 |
-1.37 |
-2.74 |
-2.74 |
2.80E-45 |
|
36 |
LMAN2 |
-1.2 |
-4.52 |
-4.49 |
1.40E-45 |
|
37 |
LSM12 |
-1.16 |
-3.31 |
-3.15 |
4.20E-44 |
|
38 |
LSM12 |
-1.17 |
-3.26 |
-3.12 |
1.40E-45 |
|
39 |
MAD2L1BP |
-1.18 |
-3.41 |
-3.11 |
2.80E-45 |
|
40 |
MALSU1 |
-1.07 |
-2.48 |
-2.41 |
6.87E-44 |
|
41 |
MAU2 |
-1.27 |
-1.88 |
-1.87 |
1.40E-45 |
|
42 |
MEA1 |
-1.36 |
-3.71 |
-3.68 |
1.54E-44 |
|
43 |
MRPL28 |
-1.38 |
-3.11 |
-3.18 |
1.40E-45 |
|
44 |
MRPL40 |
-1.07 |
-3.43 |
-3.38 |
1.40E-45 |
|
45 |
MRPS11 |
-1.24 |
-4.38 |
-4.2 |
1.40E-45 |
|
46 |
MRPS18A |
-1.6 |
-4.22 |
-4.43 |
1.40E-45 |
|
47 |
NDUFAF3 |
-1.44 |
-4.7 |
-4.49 |
1.40E-45 |
|
48 |
NELFE |
-1.11 |
-2.11 |
-2.28 |
3.78E-44 |
|
49 |
NELFE |
-1.11 |
-2.11 |
-2.28 |
2.80E-45 |
|
50 |
OR7E14P |
-1.68 |
-2.8 |
-2.89 |
1.40E-45 |
|
51 |
OR7E12P; |
-1.68 |
-2.8 |
-2.89 |
1.40E-45 |
|
52 |
OR7E26P |
-1.81 |
-3.05 |
-3.25 |
4.76E-44 |
|
53 |
FSCN3 |
-1.96 |
-3.17 |
-3.29 |
1.40E-45 |
|
54 |
OR7E12P |
-1.71 |
-2.8 |
-2.89 |
2.80E-45 |
|
55 |
OR7E14P |
-1.78 |
-2.77 |
-2.92 |
1.40E-45 |
|
56 |
TGIF2 |
-1.09 |
-3.79 |
-3.42 |
1.40E-45 |
|
57 |
OR7E37P |
-1.75 |
-2.68 |
-2.76 |
1.40E-45 |
|
58 |
OR7E12P |
-1.71 |
-2.86 |
-2.92 |
1.40E-45 |
|
59 |
OR7E14P |
-1.7 |
-2.85 |
-2.92 |
1.68E-44 |
|
60 |
HIST2H2BC |
-1.45 |
-2.97 |
-3.02 |
2.10E-44 |
|
61 |
OR7E55P |
-1.43 |
-2.21 |
-2.12 |
1.40E-45 |
|
62 |
PEBP1 |
-1.19 |
-3.91 |
-3.49 |
4.76E-44 |
|
63 |
PSMB6 |
-1.03 |
-4.42 |
-3.86 |
1.40E-45 |
|
64 |
PSMD8 |
-1.27 |
-3.11 |
-2.86 |
1.40E-45 |
|
65 |
PSMG2 |
-1.05 |
-2.48 |
-2.49 |
1.40E-45 |
|
66 |
PXN |
-1.47 |
-3.3 |
-3.19 |
1.40E-45 |
|
67 |
RAB5C |
-1.22 |
-2.84 |
-2.78 |
1.40E-45 |
|
68 |
RNU2-1; WDR74 |
-1.56 |
-5.79 |
-5.89 |
1.40E-45 |
|
69 |
RNU2-1; WDR74 |
-1.47 |
-5.39 |
-5.25 |
3.08E-44 |
|
70 |
RNU2-1; WDR74 |
-1.42 |
-5.13 |
-5.01 |
1.40E-45 |
|
71 |
RPL27A |
-1.13 |
-16.61 |
-16.21 |
3.36E-44 |
|
72 |
SNORD116@ |
-1.59 |
-6.69 |
-6.94 |
1.40E-45 |
|
73 |
SNORD13P3 |
-1.24 |
-8.33 |
-9.2 |
1.40E-45 |
|
74 |
SNHG1 |
-1.39 |
-9.65 |
-9.93 |
1.40E-45 |
|
75 |
RABGGTB |
-1.64 |
-19.64 |
-20.13 |
1.40E-45 |
|
76 |
NOP56 |
-1.3 |
-3.79 |
-4.17 |
1.40E-45 |
|
77 |
COX16 |
-1.03 |
-3.33 |
-3.21 |
1.40E-45 |
|
78 |
SNHG12 |
-1.18 |
-7.14 |
-6.36 |
4.34E-44 |
|
79 |
RNU4-1 |
-1.64 |
-11.58 |
-10.52 |
1.40E-45 |
|
80 |
RNU4-2 |
-1.09 |
-13.03 |
-17.19 |
1.40E-45 |
|
81 |
RNU4ATAC |
-1.61 |
-17.27 |
-15.58 |
1.96E-44 |
|
82 |
RNVU1-18 |
-1.82 |
-9.15 |
-8.04 |
1.40E-45 |
|
83 |
RNU1-3 |
-1.82 |
-9.15 |
-8.04 |
1.40E-44 |
|
84 |
RNU1-4 |
-1.81 |
-8.37 |
-7.42 |
1.40E-45 |
|
85 |
RNU1-2 |
-1.81 |
-8.37 |
-7.42 |
1.40E-45 |
|
86 |
RNU1-1 |
-1.81 |
-8.37 |
-7.42 |
4.06E-44 |
|
87 |
RNU1-28P; |
-1.81 |
-8.37 |
-7.42 |
1.40E-45 |
|
88 |
RNU1-27P |
-1.81 |
-8.37 |
-7.42 |
1.82E-44 |
|
89 |
RNU1-27P |
-1.8 |
-8.26 |
-7.33 |
1.40E-45 |
|
90 |
RPL23AP5 |
-1 |
-2.22 |
-2.28 |
1.40E-45 |
|
91 |
RPL18A |
-1.73 |
-3.29 |
-3.41 |
1.40E-45 |
|
92 |
RPL23A |
-1 |
-2.22 |
-2.28 |
1.40E-45 |
|
93 |
RPL23A |
-1 |
-2.32 |
-2.34 |
4.76E-44 |
|
94 |
RPL36 |
-1.19 |
-3.24 |
-3.22 |
5.61E-45 |
|
95 |
SCARNA4 |
-8.4 |
-20.56 |
-20.51 |
1.40E-45 |
|
96 |
SCYL1 |
-1.25 |
-2.33 |
-2.29 |
1.26E-44 |
|
97 |
SELPLG |
-1.57 |
-6.29 |
-5.76 |
6.03E-44 |
|
98 |
SF3B5 |
-1.56 |
-3.65 |
-3.61 |
1.68E-44 |
|
99 |
SKI |
-1.44 |
-2.88 |
-2.75 |
1.40E-45 |
|
100 |
SLC25A6 |
-1.25 |
-3.88 |
-3.73 |
1.40E-45 |
|
101 |
SLC25A6 |
-1.12 |
-3.4 |
-3.24 |
1.40E-45 |
|
102 |
SLC25A6 |
-1.12 |
-3.4 |
-3.24 |
8.41E-45 |
|
103 |
SNORA16A |
-1.18 |
-7.14 |
-6.36 |
4.34E-44 |
|
104 |
SNORA20 |
-1.79 |
-9.5 |
-9.15 |
2.80E-45 |
|
105 |
SNORA22 |
-2.05 |
-8.32 |
-11.01 |
1.40E-45 |
|
106 |
SNORA23 |
-1.59 |
-10.99 |
-10.18 |
1.40E-45 |
|
107 |
SNORA38B |
-1.82 |
-9.1 |
-8.16 |
2.52E-44 |
|
108 |
SNORA3A |
-1.13 |
-16.61 |
-16.21 |
3.36E-44 |
|
109 |
SNORA60 |
-2.96 |
-14.61 |
-15.23 |
1.40E-45 |
|
110 |
SNORA71D |
-1.39 |
-16.4 |
-15.62 |
1.40E-45 |
|
111 |
SNORD116-14 |
-1.59 |
-6.19 |
-6.1 |
1.40E-45 |
|
112 |
SNORD116-15 |
-1.27 |
-4.73 |
-4.96 |
1.40E-45 |
|
113 |
SNORD116-20 |
-1.59 |
-6.69 |
-6.94 |
1.40E-45 |
|
114 |
SNORD116-23 |
-1.75 |
-6.72 |
-6.61 |
1.40E-45 |
|
115 |
SNORD116-24 |
-1.58 |
-7.39 |
-7.98 |
1.40E-45 |
|
116 |
SNORD13 |
-1.24 |
-8.33 |
-9.2 |
1.40E-45 |
|
117 |
SNORD13P3 |
-1.39 |
-9.65 |
-9.93 |
1.40E-45 |
|
118 |
SNORD29 |
-1.22 |
-4.77 |
-4.89 |
1.40E-45 |
|
119 |
SNORD32B |
-1.38 |
-3.22 |
-3.07 |
8.41E-45 |
|
120 |
SNORD41 |
-1.64 |
-19.64 |
-20.13 |
1.40E-45 |
|
121 |
SNORD45A |
-1.59 |
-4.98 |
-5.21 |
1.40E-45 |
|
122 |
SNORD57 |
-1.3 |
-3.79 |
-4.17 |
1.40E-45 |
|
123 |
SYNJ2BP- |
-1.03 |
-3.33 |
-3.21 |
1.40E-45 |
|
124 |
TMED4 |
-1.42 |
-3.08 |
-3.13 |
7.01E-45 |
|
125 |
TMEM14C |
-1.31 |
-3.1 |
-2.94 |
1.40E-45 |
|
126 |
TMEM160 |
-1.47 |
-3.05 |
-3.12 |
1.40E-45 |
|
127 |
TMEM179B |
-1.53 |
-4.35 |
-4.4 |
1.40E-45 |
|
128 |
TMEM248 |
-1.21 |
-2.67 |
-2.65 |
2.80E-45 |
|
129 |
TMEM261 |
-1.41 |
-4.49 |
-4.37 |
1.40E-45 |
|
130 |
TMEM50A |
-1.21 |
-2.37 |
-2.41 |
1.40E-45 |
|
131 |
TMEM53 |
-1.66 |
-3.64 |
-3.81 |
1.40E-45 |
|
132 |
TPST2 |
-1.5 |
-3.01 |
-2.84 |
1.40E-45 |
|
133 |
TRIM39-RPP21; RPP21 |
-1.58 |
-4.66 |
-4.53 |
1.40E-45 |
|
134 |
TRIM39-RPP21; RPP21 |
-1.58 |
-4.66 |
-4.53 |
1.40E-45 |
|
135 |
TRIM39-RPP21; RPP21 |
-1.57 |
-4.11 |
-3.94 |
7.01E-45 |
|
136 |
UBE2A |
-1.06 |
-2.11 |
-2.16 |
1.40E-45 |
|
137 |
UBE2Q1 |
-1.08 |
-1.99 |
-1.96 |
1.40E-45 |
|
138 |
UBE2R2 |
-1.18 |
-2.34 |
-2.3 |
9.81E-44 |
|
139 |
UCKL1 |
-1.37 |
-2.4 |
-2.38 |
1.40E-45 |
|
140 |
UFC1 |
-1.11 |
-3.49 |
-4 |
2.80E-44 |
|
141 |
URGCP-MRPS24; MRPS24 |
-1.34 |
-4.26 |
-4.2 |
8.41E-45 |
|
142 |
VTRNA1-1 |
-2.5 |
-51.35 |
-49.39 |
1.40E-45 |
|
143 |
WFS1 |
-2.04 |
-4 |
-4.08 |
8.41E-45 |
|
144 |
XRCC1 |
-1.47 |
-2.63 |
-2.79 |
2.24E-44 |
|
145 |
ZNF384 |
-1.16 |
-1.72 |
-1.74 |
1.40E-45 |
Table 3(i): 145 DEGs commonly downregulated gene out of total 353 in PAH, PF with PH and PF with no PH compared to healthy control with condition FDR F<1E-43.
|
S.no. |
Gene Symbol |
A vs C Fold Change |
B vs C Fold Change |
D vs C Fold Change |
Condition FDR F-Test |
|
1 |
ABCD3 |
1.78 |
5.36 |
4.82 |
1.40E-45 |
|
2 |
ABI1 |
1.25 |
2.3 |
2.24 |
1.40E-45 |
|
3 |
ADSS |
1.7 |
3.16 |
2.99 |
1.40E-45 |
|
4 |
AGPS |
2 |
4.41 |
4.36 |
1.40E-45 |
|
5 |
ANP32E |
1.7 |
3.86 |
3.9 |
1.40E-45 |
|
6 |
ARMCX3 |
1.46 |
4.8 |
4.86 |
1.40E-44 |
|
7 |
ATL2 |
1.54 |
5.81 |
5.11 |
1.40E-44 |
|
8 |
ATP6V1C1 |
2.16 |
4.16 |
4.12 |
1.40E-44 |
|
9 |
ATRX |
2.66 |
6.58 |
6.18 |
1.40E-45 |
|
10 |
AZIN1 |
1.64 |
4.34 |
4.06 |
1.40E-45 |
|
11 |
B3GALNT2 |
1.29 |
2.99 |
2.78 |
1.40E-45 |
|
12 |
BMS1 |
1.8 |
4.28 |
4.14 |
1.40E-45 |
|
13 |
BNIP2 |
1.7 |
3.13 |
3.15 |
1.40E-45 |
|
14 |
BRMS1L |
1.72 |
4.23 |
4.26 |
1.40E-45 |
|
15 |
BZW1 |
2.34 |
3.86 |
3.85 |
1.40E-45 |
|
16 |
C11orf58 |
1.56 |
2.56 |
2.36 |
5.61E-45 |
|
17 |
C6orf62 |
1.62 |
2.99 |
2.94 |
8.41E-45 |
|
18 |
CAAP1 |
1.38 |
2.97 |
2.84 |
2.80E-44 |
|
19 |
CCDC186; MIR2110 |
3.84 |
6.89 |
6.22 |
1.40E-45 |
|
20 |
CCDC47 |
2.07 |
3.51 |
3.59 |
1.68E-44 |
|
21 |
CCDC82 |
1.6 |
3.69 |
3.72 |
7.01E-45 |
|
22 |
CEP290 |
2.47 |
5.25 |
4.81 |
8.41E-45 |
|
23 |
CLCN3 |
1.66 |
3.79 |
3.85 |
1.40E-45 |
|
24 |
CLPX |
1.77 |
3.48 |
3.37 |
1.54E-44 |
|
25 |
CNBP |
1.31 |
2.15 |
2.18 |
1.40E-45 |
|
26 |
COL4A3BP |
1.81 |
3.38 |
3.04 |
1.40E-45 |
|
27 |
COPB1 |
2.7 |
3.58 |
3.65 |
1.40E-45 |
|
28 |
CSNK1A1 |
1.59 |
3.72 |
3.86 |
1.40E-45 |
|
29 |
CTR9 |
2.53 |
5.67 |
5.58 |
1.40E-45 |
|
30 |
CWC27 |
2.06 |
3.26 |
3.26 |
7.01E-44 |
|
31 |
DCUN1D1 |
1.47 |
3.57 |
3.6 |
1.40E-45 |
|
32 |
DDX3X |
2.38 |
4.58 |
4.73 |
1.40E-45 |
|
33 |
DDX42 |
1.43 |
3.78 |
3.66 |
1.40E-45 |
|
34 |
DDX46 |
2.04 |
3.34 |
3.16 |
1.40E-45 |
|
35 |
DDX50 |
1.2 |
3.38 |
3.15 |
1.40E-45 |
|
36 |
DEK |
2.04 |
3.92 |
3.6 |
1.40E-45 |
|
37 |
DLD |
2.33 |
4.71 |
4.58 |
4.20E-44 |
|
38 |
DNAJA2 |
1.53 |
3.77 |
3.61 |
1.40E-45 |
|
39 |
DNAJC10 |
2.19 |
5.17 |
5.42 |
2.52E-44 |
|
40 |
DNAJC3 |
2.65 |
4.28 |
4.81 |
1.40E-45 |
|
41 |
EID1 |
1.15 |
3.2 |
2.92 |
1.40E-45 |
|
42 |
EIF4A2 |
1.81 |
6.18 |
6.67 |
1.40E-45 |
|
43 |
EIF5B |
2.75 |
4.93 |
5.08 |
1.40E-45 |
|
44 |
ENOPH1 |
1.19 |
3.22 |
3.28 |
1.40E-45 |
|
45 |
EPRS |
3.3 |
6.55 |
6.68 |
1.40E-45 |
|
46 |
ESF1 |
2.25 |
7.17 |
6.95 |
1.40E-45 |
|
47 |
ETNK1 |
1.42 |
3.85 |
3.45 |
7.01E-45 |
|
48 |
EWSR1 |
1.01 |
2.87 |
2.96 |
2.80E-45 |
|
49 |
EXOC5 |
2.28 |
3.73 |
3.72 |
4.20E-45 |
|
50 |
FAM133B; FAM133DP |
2.27 |
4.15 |
4.3 |
1.40E-45 |
|
51 |
FAM133DP; FAM133B |
2.32 |
4.05 |
4.08 |
1.40E-45 |
|
52 |
FAM133DP; FAM133B |
2.41 |
4.49 |
4.53 |
3.50E-44 |
|
53 |
FAM208A |
1.36 |
3.14 |
3.08 |
1.68E-44 |
|
54 |
FAM3C |
1.56 |
4.46 |
4.5 |
2.80E-45 |
|
55 |
FAM3C; FAM3C2 |
1.59 |
4.46 |
4.46 |
5.61E-44 |
|
56 |
FAR1 |
1.67 |
3.7 |
3.52 |
3.78E-44 |
|
57 |
FBXO11 |
1.81 |
3.56 |
3.52 |
2.80E-45 |
|
58 |
FBXO28 |
1.67 |
2.58 |
2.52 |
1.40E-45 |
|
59 |
FGFR1OP2 |
1.49 |
3.3 |
3.53 |
1.96E-44 |
|
60 |
FKBP3 |
1.12 |
2.66 |
2.59 |
9.25E-44 |
|
61 |
FXR1 |
2.38 |
6.17 |
5.76 |
1.40E-45 |
|
62 |
FYTTD1 |
1.65 |
2.87 |
2.96 |
1.40E-45 |
|
63 |
GBE1 |
2.11 |
3.6 |
3.7 |
1.40E-45 |
|
64 |
GCC2 |
3.48 |
7.6 |
6.91 |
1.40E-45 |
|
65 |
GGNBP2 |
2.07 |
3.63 |
3.56 |
1.40E-45 |
|
66 |
GLOD4 |
1.06 |
2.87 |
2.81 |
1.40E-45 |
|
67 |
GNAI3 |
1.7 |
2.91 |
3.13 |
1.40E-45 |
|
68 |
GOLGA4 |
2.86 |
4.12 |
4.25 |
1.40E-45 |
|
69 |
GOLGA6L17 |
1.35 |
3.94 |
3.5 |
1.40E-45 |
|
70 |
GOLGA6L9 |
1.31 |
4.03 |
3.6 |
1.40E-45 |
|
71 |
GOLGB1 |
2.73 |
5.13 |
5.08 |
1.40E-45 |
|
72 |
GOLT1B |
1.66 |
4.72 |
4.98 |
1.40E-45 |
|
73 |
GTF3C3 |
2.11 |
3.93 |
3.89 |
1.40E-45 |
|
74 |
HERC4 |
1.53 |
3.78 |
3.44 |
1.40E-45 |
|
75 |
HNRNPA1P10 |
2.11 |
3.88 |
3.82 |
7.01E-45 |
|
76 |
HNRNPA1P1 |
2.27 |
4.07 |
3.93 |
1.40E-45 |
|
77 |
HNRNPA3 |
2.37 |
5.28 |
4.83 |
1.40E-45 |
|
78 |
HNRNPA3 |
2.37 |
5.39 |
4.88 |
1.40E-45 |
|
79 |
HNRNPH1 |
1.6 |
4.1 |
4.16 |
1.40E-45 |
|
80 |
HNRNPH2; RPL36A-HNRNPH2 |
1.59 |
2.96 |
3 |
1.40E-45 |
|
81 |
HNRNPM |
1.26 |
3.04 |
3.03 |
1.40E-45 |
|
82 |
HNRNPR |
1.75 |
3.38 |
3.35 |
1.40E-45 |
|
83 |
HNRNPU |
1.67 |
3.39 |
3.41 |
1.40E-45 |
|
84 |
HS2ST1 |
1.47 |
2.65 |
2.66 |
1.40E-45 |
|
85 |
HTATSF1 |
1.63 |
5.02 |
4.5 |
1.40E-45 |
|
86 |
IFT80 |
1.92 |
6.11 |
5.54 |
8.41E-45 |
|
87 |
INSIG2 |
1.25 |
5.06 |
4.82 |
2.80E-45 |
|
88 |
ITCH |
1.43 |
2.35 |
2.4 |
1.40E-45 |
|
89 |
KTN1 |
3.08 |
6.1 |
5.57 |
1.40E-45 |
|
90 |
LBR |
1.19 |
3.06 |
3.2 |
1.40E-45 |
|
91 |
LEO1 |
1.84 |
4.59 |
4.2 |
1.40E-45 |
|
92 |
LRPPRC |
2.78 |
5 |
4.99 |
1.96E-44 |
|
93 |
LUC7L3 |
2.14 |
6.7 |
6.27 |
1.40E-45 |
|
94 |
MED4 |
1.64 |
2.94 |
2.64 |
1.40E-45 |
|
95 |
MFN1 |
1.97 |
3.78 |
3.6 |
1.40E-45 |
|
96 |
MIB1 |
1.62 |
3.8 |
3.42 |
5.61E-45 |
|
97 |
MIER1 |
1.61 |
2.63 |
2.76 |
1.40E-45 |
|
98 |
MOB1A |
1.25 |
2.59 |
2.6 |
1.40E-45 |
|
99 |
MPHOSPH10 |
2.15 |
5.84 |
5.58 |
5.61E-45 |
|
100 |
MRFAP1 |
1.2 |
2.55 |
2.53 |
1.40E-45 |
|
101 |
MRPL1 |
1.59 |
3.47 |
3.47 |
1.40E-45 |
|
102 |
MSANTD4 |
2.35 |
5.11 |
4.73 |
8.55E-44 |
|
103 |
NAP1L1 |
1.82 |
3.01 |
2.92 |
1.40E-45 |
|
104 |
NBPF20 |
1.89 |
2.82 |
2.78 |
7.01E-45 |
|
105 |
NBPF1 |
1.97 |
3.1 |
3.05 |
2.80E-45 |
|
106 |
NBPF14 |
2.03 |
3.08 |
3.02 |
2.80E-45 |
|
107 |
NBPF14 |
1.93 |
2.9 |
2.84 |
1.40E-45 |
|
108 |
NDUFA5 |
1.17 |
4.92 |
3.84 |
1.40E-45 |
|
109 |
NEMF |
2.03 |
3.25 |
3.34 |
1.40E-45 |
|
110 |
NFYB |
1.05 |
4.49 |
4.4 |
1.40E-45 |
|
111 |
NMD3 |
1.85 |
4.87 |
4.9 |
1.40E-45 |
|
112 |
NUP107 |
1.76 |
4.82 |
4.98 |
2.80E-45 |
|
113 |
OPA1 |
2.68 |
4.85 |
4.81 |
1.40E-45 |
|
114 |
PAFAH1B1 |
1.42 |
2.73 |
2.66 |
1.40E-45 |
|
115 |
PDIA3 |
1.9 |
3.65 |
3.53 |
1.40E-45 |
|
116 |
PDIA3 |
1.88 |
4.32 |
4.11 |
1.40E-45 |
|
117 |
PDIA6 |
1.89 |
4.64 |
4.68 |
1.40E-45 |
|
118 |
PHF14 |
1.92 |
3.4 |
3.25 |
1.40E-45 |
|
119 |
PI4K2B |
1.51 |
3.52 |
3.31 |
1.40E-45 |
|
120 |
PITPNB |
1.68 |
3.99 |
4.05 |
1.40E-45 |
|
121 |
PLRG1 |
1.97 |
4.58 |
4.65 |
1.40E-45 |
|
122 |
PNN |
2.17 |
6.05 |
5.67 |
1.40E-45 |
|
123 |
POLR2B |
2.23 |
5.36 |
5.43 |
1.40E-45 |
|
124 |
PPP3R1 |
1.34 |
1.97 |
1.96 |
1.40E-45 |
|
125 |
PPP4R2 |
2.17 |
4.78 |
4.54 |
1.40E-45 |
|
126 |
PPP4R3A |
1.52 |
2.33 |
2.4 |
1.40E-45 |
|
127 |
PRKAR1A |
1.29 |
2.1 |
1.97 |
1.40E-45 |
|
128 |
PRPF38B |
1.71 |
2.94 |
3.06 |
1.40E-45 |
|
129 |
PRPF39 |
1.6 |
4.02 |
3.89 |
1.40E-44 |
|
130 |
RAB18 |
1.84 |
3.24 |
3.11 |
1.40E-45 |
|
131 |
RAB1A |
1.8 |
3.37 |
3.38 |
1.40E-45 |
|
132 |
RABEP1 |
2.25 |
3.29 |
3.3 |
5.61E-45 |
|
133 |
RABGGTB; ACADM |
1.63 |
3.36 |
3.27 |
1.40E-45 |
|
134 |
RAD21 |
2.04 |
4.31 |
4.07 |
2.80E-45 |
|
135 |
RANBP2 |
2.04 |
4.04 |
4.03 |
1.40E-45 |
|
136 |
RB1CC1 |
2.21 |
4.39 |
4.2 |
2.80E-44 |
|
137 |
RBM34; ARID4B |
1.48 |
2.89 |
3.15 |
1.40E-45 |
|
138 |
RCHY1 |
1.57 |
3.52 |
3.38 |
1.40E-45 |
|
139 |
RECQL |
2.15 |
4 |
4.14 |
9.81E-45 |
|
140 |
RNF219 |
1.99 |
4.86 |
5.22 |
1.40E-45 |
|
141 |
RNF6 |
2.56 |
4.01 |
3.87 |
9.25E-44 |
|
142 |
RNMT |
2.02 |
4.42 |
4.47 |
1.40E-45 |
|
143 |
ROCK1 |
3.18 |
4.77 |
4.68 |
8.13E-44 |
|
144 |
RSF1 |
2.23 |
4.78 |
4.3 |
1.40E-45 |
|
145 |
RSL24D1 |
1.98 |
3.35 |
3.12 |
1.40E-45 |
|
146 |
SBNO1 |
2.27 |
5.62 |
5.34 |
5.89E-44 |
|
147 |
SEC23IP |
1.54 |
3.52 |
3.7 |
1.40E-45 |
|
148 |
SEC62 |
1.75 |
3.25 |
3.06 |
1.40E-45 |
|
149 |
SESN3 |
1.67 |
5.69 |
5.22 |
1.26E-44 |
|
150 |
SKIV2L2 |
2.24 |
4.62 |
4.79 |
8.41E-45 |
|
151 |
SLC33A1 |
1.25 |
3.34 |
3.27 |
2.80E-45 |
|
152 |
SLC35A3 |
1.16 |
3.34 |
3.17 |
1.40E-45 |
|
153 |
SLC35D1 |
1.27 |
3.88 |
3.81 |
1.40E-45 |
|
154 |
SLTM |
2.49 |
4.31 |
4.26 |
1.40E-45 |
|
155 |
SLU7 |
3.09 |
6.21 |
6.25 |
1.40E-45 |
|
156 |
SMARCA5 |
2.04 |
3.87 |
3.78 |
9.81E-45 |
|
157 |
SNX4 |
1.77 |
3.98 |
3.68 |
1.40E-45 |
|
158 |
SREK1 |
2.31 |
4.79 |
4.68 |
1.40E-45 |
|
159 |
SRSF10 |
1.79 |
3.21 |
3.17 |
1.40E-45 |
|
160 |
SRSF4 |
1.67 |
2.82 |
2.81 |
1.40E-45 |
|
161 |
STAG1 |
2.06 |
4.02 |
3.87 |
1.40E-45 |
|
162 |
STXBP3 |
1.67 |
3.02 |
2.86 |
3.08E-44 |
|
163 |
SYF2 |
1.73 |
3.56 |
3.41 |
1.40E-45 |
|
164 |
TAX1BP1 |
2.83 |
4.29 |
4.03 |
1.40E-45 |
|
165 |
TBC1D23 |
1.96 |
2.58 |
2.65 |
1.40E-45 |
|
166 |
TCEA1 |
1.74 |
3.15 |
2.88 |
1.40E-45 |
|
167 |
TCEA1 |
1.85 |
3.66 |
3.29 |
8.41E-45 |
|
168 |
THUMPD1 |
2 |
3.48 |
3.29 |
1.40E-45 |
|
169 |
TM9SF3 |
1.58 |
2.71 |
2.64 |
2.94E-44 |
|
170 |
TMEM167A |
1.62 |
3.47 |
3.1 |
4.20E-45 |
|
171 |
TMF1 |
2.77 |
7.61 |
7.81 |
1.40E-45 |
|
172 |
TMX3 |
1.62 |
3.68 |
3.66 |
5.47E-44 |
|
173 |
TOP2B |
2.41 |
5.69 |
5.1 |
1.40E-45 |
|
174 |
TPR |
3.18 |
4.85 |
4.73 |
1.40E-45 |
|
175 |
TRAM1 |
1.28 |
3.07 |
3 |
1.40E-45 |
|
176 |
TRAPPC8 |
1.81 |
4.02 |
4.01 |
1.40E-45 |
|
177 |
TSN |
1.42 |
2.42 |
2.32 |
1.40E-45 |
|
178 |
TSPAN3 |
1.23 |
2.63 |
2.53 |
1.40E-45 |
|
179 |
TTC3 |
2.55 |
4.71 |
4.32 |
1.40E-45 |
|
180 |
TVP23B |
1.56 |
3.01 |
2.75 |
1.40E-45 |
|
181 |
TYW3 |
1.79 |
3.54 |
3.49 |
1.40E-45 |
|
182 |
UBA5 |
1.82 |
3.31 |
3.07 |
5.61E-45 |
|
183 |
UBE2V2 |
1.29 |
2.74 |
2.59 |
1.40E-45 |
|
184 |
UBE3A |
1.46 |
2.95 |
2.88 |
2.66E-44 |
|
185 |
UBXN4 |
2.32 |
3.69 |
3.65 |
1.12E-44 |
|
186 |
UHMK1 |
1.31 |
3.55 |
3.43 |
1.40E-45 |
|
187 |
UHRF1BP1L |
2.26 |
2.95 |
2.94 |
1.40E-45 |
|
188 |
USP1 |
1.89 |
3.59 |
3.34 |
9.81E-45 |
|
189 |
USP16 |
2.92 |
3.41 |
3.55 |
1.40E-45 |
|
190 |
USP33 |
1.32 |
3.62 |
3.51 |
8.69E-44 |
|
191 |
USP47 |
2.29 |
6.47 |
6.4 |
4.20E-45 |
|
192 |
VPS4B |
1.56 |
3.02 |
2.84 |
1.40E-45 |
|
193 |
WDR35 |
1.64 |
4.4 |
4.13 |
4.76E-44 |
|
194 |
YTHDC1 |
1.89 |
3.3 |
3.29 |
1.40E-45 |
|
195 |
ZC3H13 |
2.7 |
4.25 |
4.4 |
8.41E-45 |
|
196 |
ZFYVE16 |
2.05 |
3.47 |
3.38 |
7.43E-44 |
|
197 |
ZNF23 |
1.11 |
2.54 |
2.55 |
9.67E-44 |
|
198 |
ZNF841 |
2.28 |
6.81 |
7.29 |
1.40E-45 |
Table 3(ii): 198 DEGs. commonly upregulated genes out of total 353 in PAH, PF with PH and PF with no PH compared to healthy control with condition FDR F<1E-43.
|
S.NO. |
Gene Symbol |
A vs C Fold Change |
B vs C Fold Change |
D vs C Fold Change |
|
1 |
ATMIN |
1.34 |
-1.84 |
-1.88 |
|
2 |
MAP1LC3B2 |
1.34 |
-2.36 |
-2.28 |
|
3 |
POMP |
1.04 |
-3.11 |
-2.89 |
|
4 |
PPP6C |
1.22 |
-1.66 |
-1.59 |
|
5 |
PTMAP3 |
1.07 |
-1.66 |
-1.64 |
|
6 |
CDK5RAP3 |
-1.03 |
3.06 |
2.86 |
|
7 |
CREBZF |
-1.12 |
2.69 |
2.43 |
|
8 |
ND6 |
-1.11 |
11.08 |
10.05 |
|
9 |
SCARNA17 |
-1.26 |
3.51 |
3.55 |
|
10 |
SOD1 |
-1.11 |
2.07 |
1.94 |
Table 4: Differentially up and down regulated 10 significant genes in PAH with respect to PF with and without PH base line control with condition FDR F test value F<1.40E-45.
3.6 DEGs in PAH and PF (with PH and PF without PH)
On the basis of fold change as shown in the heatmap (Figure 6) top 5 DEGs among the total commonly downregulated in PAH and PF both were namely (OR7E12P, VTRNA1.1, SELPLG, SKI, SNORDA20) and among the commonly upregulated, top 5 DEGs were (LEO1, RNMT, GOLT1B, NMD3, GOLGAH) as shown in (Figure 7). Whereas, PAH showed differential regulation in 10 DEGs) (Figure 8) on comparison of PAH and PF (with PH and PF without PH) including 5 upregulated DEGs. in PAH (PTMAP3, PPP6C, ATMIN, MAPILC, POMP) and 5 downregulated DEGs. in PAH (CREBZF, CDK5RA, SOD1,SCARNA, ND6. Differentially regulated top 5 upregulated DEGs in PAH were plotted on log2 gene expression level to observe the intensities of each gene expression (Figure 9) (Table 5).
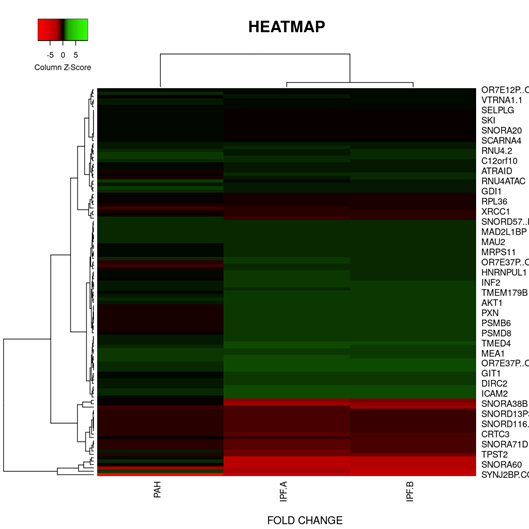
Figure 6: Heat map and dendogram shows, Hierarchical Cluster analysis of the top 39 DEGs. commonly downregulated in both groups (PAH and PF) out of 353 total DEGs. ,with FDR condition F test ( F< 1E-43). Distance metric used between objects is the Euclidean distance. Distances between clusters of objects are computed using the complete linkage method.
PAH*- PAH, PF-A*- PF WITH PH , PF-B*- PF WITH NO PH
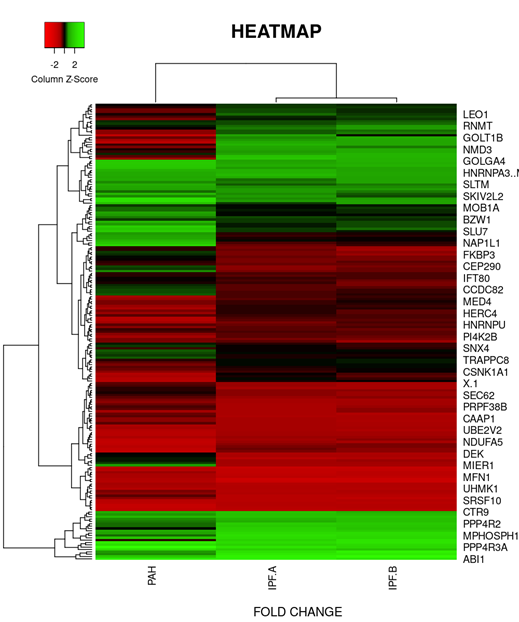
Figure 7: Heat map and dendogram shows, hierarchical cluster analysis of the top 39 DEGs. commonly upregulated in both groups (PAH and IPF) gene out of 353 with FDR condition F test (F< 1E-43). Distance metric used between objects is the Euclidean distance. Distances between clusters of objects are computed using the complete linkage method.
PAH*- PAH, PF-A*- PF WITH PH , PF-B*- PF WITH NO PH
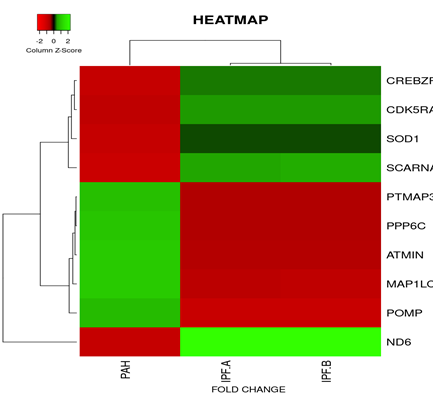
Figure 8: Heat map and dendogram shows, hierarchical cluster analysis of the top 10 Differentially regulated DEGs. in PAH out of 353 genes with FDR condition F test (F < 1E-43). Distance metric used between objects is the Euclidean distance. Distances between clusters of objects are computed using the complete linkage method.
PAH*- PAH, PF-A*- PF WITH PH , PF-B*- PF WITH NO PH
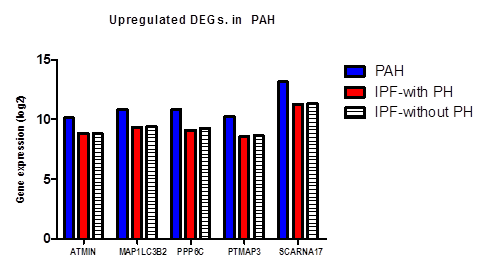
Figure 9: Gene expression level in log2 manner for 5 differentially upregulated genes in PAH.
|
Gene Symbol |
PAH(log2) |
IPF-A(log2) |
IPF-B(log2) |
|
ATMIN |
10.13 |
8.84 |
8.81 |
|
CDK5RAP3 |
8.53 |
7.29 |
7.42 |
|
CREBZF |
10.05 |
7.81 |
7.93 |
|
MAP1LC3B2 |
10.82 |
9.34 |
9.4 |
|
ND6 |
10.46 |
11.19 |
11.14 |
|
POMP |
9.87 |
10.81 |
10.77 |
|
PPP6C |
10.82 |
9.1 |
9.27 |
|
PTMAP3 |
10.24 |
8.55 |
8.65 |
|
SCARNA17 |
13.13 |
11.28 |
11.31 |
|
SOD1 |
9.78 |
10.82 |
10.88 |
Table 5: Differentially up regulated 10 significant genes expression (log 2) in PAH with respect to PF with and without PH base line control with with condition FDR F<1E-43.
3.7 Pathway and gene ontology associated with PAH and PF (with PH and PF without PH)
On submitting a total of 343 DEGs commonly regulated in PAH and PF, REACTOME gave 48 significant pathways () showing involvement of 14 genes (Table 6). And among 10 differentially expressed genes in PAH, 3 genes were respectively involved in 6 pathways such as (Interleukin-12 signaling, Complex I biogenesis etc) (Table 7). Gene ontology by PANTHER for 10 differentially regulated genes gave its associated biological processes (such as cellular component biogenesis, metabolic processes, biological regulation ) (Figure 10).
|
s.no. |
Submitted entities found |
Pathway name |
Entities pValue |
|
1 |
ABI1;AKT1;BRK1 |
VEGFA-VEGFR2 Pathway |
2.05E-04 |
|
2 |
ABI1;AKT1;BRK1 |
Signaling by VEGF |
2.82E-04 |
|
3 |
CEBPD;AKT1 |
Interleukin-4 and Interleukin-13 signaling |
0.001405 |
|
4 |
AKT1 |
CTLA4 inhibitory signaling |
0.001571 |
|
5 |
AKT1 |
CD28 dependent PI3K/Akt signaling |
0.001696 |
|
6 |
CCND3;CEBPD |
Transcriptional regulation of white adipocyte differentiation |
0.002012 |
|
7 |
AKT1 |
G beta:gamma signalling through PI3Kgamma |
0.002101 |
|
8 |
AKT1 |
Constitutive Signaling by AKT1 E17K in Cancer |
0.002547 |
|
9 |
AKT1 |
CD28 co-stimulation |
0.003745 |
|
10 |
AKT1 |
G-protein beta:gamma signalling |
0.003745 |
|
11 |
ABI1;BRK1 |
RHO GTPases Activate WASPs and WAVEs |
0.004127 |
|
12 |
AKT1 |
VEGFR2 mediated vascular permeability |
0.004733 |
|
13 |
CEBPD |
Defective SLC24A1 causes congenital stationary night blindness 1D (CSNB1D) |
0.009227 |
|
14 |
ABI1;AKT1;BRK1;ATP6V1C1 |
Signaling by Receptor Tyrosine Kinases |
0.013647 |
|
15 |
AKT1 |
AKT-mediated inactivation of FOXO1A |
0.01381 |
|
16 |
CEBPD;AKT1;ARF5 |
Signaling by Interleukins |
0.015185 |
|
17 |
AKT1 |
Costimulation by the CD28 family |
0.021312 |
|
18 |
ARF5 |
Nef Mediated CD4 Down-regulation |
0.022913 |
|
19 |
AKT1 |
PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 |
0.025176 |
|
20 |
ARF5 |
COPI-dependent Golgi-to-ER retrograde traffic |
0.025563 |
|
21 |
ARF5 |
COPI-mediated anterograde transport |
0.025563 |
|
22 |
AKT1 |
AKT phosphorylates targets in the nucleus |
0.027434 |
|
23 |
AKT1 |
Regulation of localization of FOXO transcription factors |
0.031935 |
|
24 |
AKT1 |
RUNX2 regulates genes involved in cell migration |
0.031935 |
|
25 |
AKT1;COX5B |
TP53 Regulates Metabolic Genes |
0.034 |
|
26 |
CEBPD |
Sodium/Calcium exchangers |
0.034178 |
|
27 |
AKT1 |
AKT phosphorylates targets in the cytosol |
0.036416 |
|
28 |
AKT1 |
Downregulation of ERBB2:ERBB3 signaling |
0.036416 |
|
29 |
AKT1 |
PI3K/AKT Signaling in Cancer |
0.038573 |
|
30 |
AKT1 |
Negative regulation of the PI3K/AKT network |
0.038573 |
|
31 |
CCND3 |
Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) |
0.038648 |
|
32 |
CCND3 |
Aberrant regulation of mitotic G1/S transition in cancer due to RB1 defects |
0.038648 |
|
33 |
AKT1 |
Regulation of TP53 Activity through Association with Co-factors |
0.038648 |
|
34 |
AKT1 |
Activation of BAD and translocation to mitochondria |
0.043099 |
|
35 |
AKT1 |
Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA |
0.043099 |
|
36 |
AKT1 |
KSRP (KHSRP) binds and destabilizes mRNA |
0.045316 |
|
37 |
CEBPD |
Activation of the phototransduction cascade |
0.045316 |
|
38 |
ARF5 |
Golgi-to-ER retrograde transport |
0.046124 |
|
39 |
COX5B;ATP5G1 |
Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
0.047245 |
|
40 |
ARF5 |
Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
0.049737 |
Table 6: Rectome pathway analysis for commonly regulated DEGs out of which 12 significant genes regulate 40 significant pathways.
|
S.NO. |
GENE NAME |
Pathway name |
Entities pValue |
|
1 |
SOD1 |
Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation |
0.001321316 |
|
2 |
SOD1 |
Interleukin-12 signaling |
0.001741671 |
|
3 |
SOD1 |
Interleukin-12 family signaling |
0.002263701 |
|
4 |
PPP6C |
Telomere Extension By Telomerase, EGF receptor signaling pathway, FGF signaling pathway (Panther) |
0.025195724 |
|
5 |
ND6 |
Complex I biogenesis |
0.041911153 |
|
6 |
SOD1 |
Detoxification of Reactive Oxygen Species |
0.047663852 |
Table 7: Rectome pathway analysis for 10 differentially regulated DEGs out of which 3 significant genes regulate 06 significant pathways.
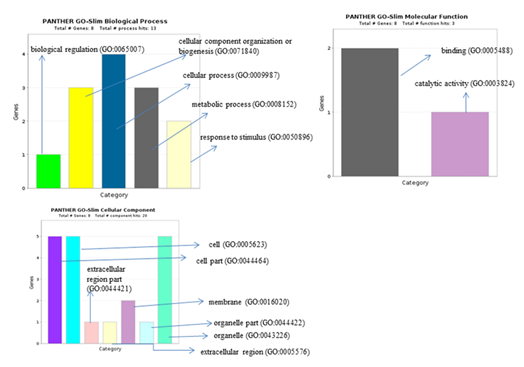
Figure 10: Panther analysis giving biological process, cellular component and molecular function of 10 differentially regulated DEGs. in PAH.
4. Discussion
The primary objective of a microarray experiment is to classify the gene expression trends in living organisms due to disease, tolerance against pathogen, a chemical compound or a certain relevant condition. For every gene, the microarray analysis tests the intensities, which means its relative degree of expression. Nevertheless, in order to
eliminate low-quality measurements, correct calculated intensities, simplification of comparisons and screening of genes that are substantially differentially expressed between samples is carried out on the data before properly associating these rates [21]. Thus, normalization is the first conversion step applied to expression data to fine-tune the individual hybridization intensities in order to be able to interpret significant biochemical associations [22].
In the present study, TAC4.0 microarray analysis revealed a total of 353 differentially expressed genes assigned with gene symbol on analyzing all the 88 lung biopsy samples with filter condition FDR F TEST (F<1E-43) within 3 groups, which are commonly or differentially regulated. 145 genes out of the total 353 get commonly downregulated and 198 gets commonly upregulated when comparing the PAH and the PF groups, proving as common markers for lung disease were as, the other 10 genes are differentially expressed in PAH between the three groups indicating group specific biomarkers. Pathway analysis through REACTOME tool for all the commonly regulated genes, with P value (P<0.05) identified 14 highly significant genes involved in 48 associated pathways and top 10 differentially regulated genes in PAH submitted to REACTOME showed output of 3 significant DEGs. involved in 06 significant pathways.
Among the top 10 differentially expressed genes, 2 (SOD1,SCARNA17) have already been reported depicting their association in idiopathic lung fibrosis and pulmonary arterial hypertension, i.e. the methyl transferase inhibitor EZH2, EPZ005687 substantially inhibits the production of TAC-induced PAH depending upon EZH2-SOD1-ROS signaling [23]. SCARNA17 (Small Cajal Body-Specific RNA 17) an RNA Gene, affiliated with the lncRNA class recently identified by microRNA expression profiling of bronchoalveolar lavage fluid cells from patients with idiopathic pulmonary fibrosis and sarcoidosis is known to be involved in IPF [24].
Going with our findings and exploring our 8 novel genetic PAH identifiers, the first one being CREBZF (CREB/ATF BZIP Transcription Factor) a coding gene is involved in multiple processes such as (negative regulation of gene expression, epigenetic modulation, negative transcription regulation, virus response, DNA-dependent regulation). Disease associated with CREBZF includes Acute Necrotizing Encephalitis. Although this gene is well explored in liver in, lipogenesis, liver regeneration and lipogenic pathway [25, 26, 27] however, we are reporting it for the first time to be involved in the genetic cause of PAH .CDK5RAP3 (CDK5 Regulatory Subunit Associated Protein 3) encodes a protein that has been reported to function in signaling pathways governing transcriptional regulation and cell cycle progression. It is known to play role in tumorigenesis and metastasis as reported in various cancers like as a tumour suppressor, CDK5RAP3 negatively controls self-renewal and invasion, and is regulated by ERK1/2 in human gastric cancer [28]. CDK5RAP3 Participates in Regulation on Autophagy and is Downregulated in Renal Cancer [29]. Lung adenocarcinoma falls under the umbrella of non-small cell lung cancer (NSCLC) and has a strong association with previous smoking. CDK5RAP3, CCNB2, and RAGE Genes are already being researched to be involved in Lung Adenocarcinoma diagnostics [30]. We thereby indicate its sole involvement in causing PAH. POMP protein (Proteasome Maturation Protein) is a molecular chaperone that binds components of 20S preproteasome, and is necessary for the creation of 20S proteasome. The 20S proteasome is the active
proteolytic portion of the 26S proteasome complex. POMP is already studied in Extracellular Alveolar Proteasome involved in Lung Injury and Repair [31] however its genetic cause in PAH is being reported by us.
ATMIN (ATM Interactor) protein plays a key role in the development of cell survival and RAD51 foci in response to damage of methylating DNA. It is majorily involved in the regulation of ATM's activity in the absence of DNA damage. ATM Signaling Network in Development and Disease are its associated pathways. Gene Ontology annotations relating to ATMIN gene includes DNA binding for the transcription regulatory region. Till now this gene is well studied in lung morphogenesis [32] and ciliogenesis, lung adenocarcinoma [33, 34], lung cancer [35], however our analysis has shown its involvement in PAH cause. MAP1LC3B2 (Microtubule Associated Protein 1 Light Chain 3 Beta 2),ubiquitin-like modifier involved in autophagosomal vacuole formation (autophagosomes). Plays a role in mitophagy that helps to control mitochondrial quantity and efficiency by removing the mitochondria at a baseline level to meet cellular energy requirements and avoid excess development of ROS. MAP1LC3B2 is studied in various studies involved in lung epithelial cell autophagy; Elastase causes autophagy of the lung epithelial cells by a placental growth factor: a new perspective into emphysema pathogenesis [36] and also in lung fibroblast studies [29]. Our results have also shown its significant expression in PAH. PTMAP3 (Prothymosin Alpha Pseudogene 3) is a pseudogene till now studied in corneal dystrophy a group of rare genetic eye disorders in which abnormal material builds up in the cornea most corneal dystrophies affect both the eyes, this progress slowly and runs in the families [37]. This gene is not much studied and our analysis gives insight to further explore this gene in being one of the genetic causes of PAH. PPP6C (Protein Phosphatase 6 Catalytic Subunit) gene encodes the protein phosphatase catalytic subunit, a component of the signalling pathway which regulates the progression of the cell cycle. PPP6C-related disorders include Pineal Parenchymal Tumor with Intermediate Differentiation and Crouzon Syndrome of Acanthosis Nigricans. Also it is studied in human glioma cells, the expression AEG-1 is correlated with levels of CD133 and PPP6c in human glioma tissue [38]. Its involvement in the PAH disease is indicated by our analysis. MT-ND6 (Mitochondrially Encoded NADH: Ubiquinone Oxidoreductase Core Subunit 6), Core subunit of NADH dehydrogenase (Complex I), the mitochondrial membrane respiratory chain, which is assumed to belong to the minimum assembly necessary for catalysis. Complex I acts for electron transport from NADH into the respiratory chain. Commonly associated diseases with MT-ND6 include Leber Optic Atrophy, Dystonia and Leber Optic Atrophy.
5. Conclusion
We proposed a method for the meta-analysis of transcriptomics studies in this article using overall effect size z score (Z=3.79), P value (P=0.0002), hetrogenity I2=43%) and confidence interval of 95%, which provides increase power for precession. Study reflects broad spectrum of PAH and its significant early biomarkers. In conclusion, a significant of 198 DEGs. commonly upregulated and 145 commonly downregulated genes with 5 up and 5 down differentially regulated DEGs. were identified from a total of 353 DEGs. including all three groups PAH, IPF with and without PH. Reactome pathway analysis for 343 commonly regulated DEGs out of total 353 gave 14 significant genes which regulate 48 significant pathways (such as-VEGFA-VEGFR2 Pathway, Interleukin-4 and Interleukin-13 signaling etc.. ) whereas, for 10 differentially regulated DEGs out of total 353, 3 genes show regulation of 06
significant pathways (such as- Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation, Interleukin-12 signaling et.). Gene ontology of 10 differentially regulated genes in PAH through Panther were involved in biological processes (such as-cellular component organization or biogenesis, metabolic pathways etc..), molecular function (such as-binding, catalytic activity) and cellular component (such as-cell and organelle etc..). Among the differential significant 10 genes a total of 2 (SOD1, SCARNA17) are already reported in PAH and IPF proving pivotal in PAH pathogenesis as indicated by our results also. (CREBZF, CDK5RAP3, POMP, ATMIN, MAP1LC3B2) genes are previously explored and reported in various lung disorders (not in PAH or IPF) but are novel identifiers in PAH as per our analysis .However, (MAP1LC3B2, PPP6C, MT-ND6) are totally novel genes obtained giving future prospects for these findings to contribute in better understanding of PAH pathogenesis, and provide a theoretical basis for further experimental studies.
Disclosure
All the authors declared no competing interests.
References
- Arcasoy SM, Christie JD, Ferrari VA, Sutton MStJ, Zisman DA, et al. Echocardiographic Assessment of Pulmonary Hypertension in Patients with Advanced Lung Disease. Am J Respir Crit Care Med 167 (2003): 735-740.
- Rubin LJ. Primary Pulmonary Hypertension. N Engl J Med 336 (1997): 111-117.
- Koh E. Pulmonary hypertension in systemic sclerosis: an analysis of 17 patients. Rheumatology 35 (1996): 989-993.
- Winslow TM, Ossipov MA, Fazio GP, Foster E, Simonson JS, et al. The left ventricle in systemic lupus erythematosus: Initial observations and a five-year follow-up in a university medical center population. Am Heart J 125 (1993): 1117-1122.
- Dawson JK. Raised pulmonary artery pressures measured with Doppler echocardiography in rheumatoid arthritis patients. Rheumatology 39 (2000): 1320-1325.
- Shorr AF, Davies DB, Nathan SD. Outcomes for Patients With Sarcoidosis Awaiting Lung Transplantation. Chest 122 (2002): 233-238.
- Shorr AF, Davies DB, Nathan SD. Predicting Mortality in Patients With Sarcoidosis Awaiting Lung Transplantation. Chest 124 (2003): 922-928.
- Crystal RG. Idiopathic Pulmonary Fibrosis: Clinical, Histologic, Radiographic, Physiologic, Scintigraphic, Cytologic, and Biochemical Aspects. Ann Intern Med 85 (1976): 769.
- Rabeyrin M, Thivolet F, Ferretti GR, Chalabreysse L, Jankowski A, et al. Usual interstitial pneumonia end-stage features from explants with radiologic and pathological correlations. Ann Diagn Pathol 19 (2015): 269-276.
- Hoeper MM, Bogaard HJ, Condliffe R, Frantz R, Khanna D, et al. Definitions and Diagnosis of Pulmonary Hypertension. J Am Coll Cardiol 62 (2013): D42-D50.
- Agrawal A, Verma I, Shah V, Agarwal A, Sikachi RR. Cardiac manifestations of idiopathic pulmonary fibrosis. Intractable Rare Dis Res 5 (2016): 70-75.
- Shorr AF, Wainright JL, Cors CS, Lettieri CJ, Nathan SD. Pulmonary hypertension in patients with pulmonary fibrosis awaiting lung transplant. Eur Respir J 30 (2007): 715-721.
- Hansdottir S, Groskreutz DJ, Gehlbach BK. WHO’s in second?: A practical review of World Health Organization group 2 pulmonary hypertension. Chest 144 (2013): 638-650.
- Staff PHA. Types of Pulmonary Hypertension: The WHO Groups [Internet]. Pulmonary Hypertension Association (2020).
- Simonneau G, Gatzoulis MA, Adatia I, Celermajer D, Denton C, et al. Updated Clinical Classification of Pulmonary Hypertension. J Am Coll Cardiol 62 (2013): D34-D41.
- Zhao YD, Chu L, Lin K, Granton E, Yin L, et al. A Biochemical Approach to Understand the Pathogenesis of Advanced Pulmonary Arterial Hypertension: Metabolomic Profiles of Arginine, Sphingosine-1-Phosphate, and Heme of Human Lung. Su X, editor. PLOS ONE 10 (2015): e0134958.
- Zhao Y, Peng J, Lu C, Hsin M, Mura M, et al. Metabolomic Heterogeneity of Pulmonary Arterial Hypertension. Sen U, editor. PLoS ONE 9 (2014): e88727.
- Zhao YD, Yun HZH, Peng J, Yin L, Chu L, et al. De novo synthesize of bile acids in pulmonary arterial hypertension lung. Metabolomics 10 (2014): 1169-1175.
- Lockstone HE, Sanderson S, Kulakova N, Baban D, Leonard A, et al. Gene Set Analysis of Lung Samples Provides Insight into Pathogenesis of Progressive, Fibrotic Pulmonary Sarcoidosis. Am J Respir Crit Care Med 181 (2010): 1367-1375.
- Baelde HJ, Eikmans M, Doran PP, Lappin DWP, de Heer E, et al. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am J Kidney Dis 43 (2004): 636-650.
- Sterne JAC, Sutton AJ, Ioannidis JPA, Terrin N, Jones DR, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ (2011).
- Leung YF, Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet 19 (2003): 649-659.
- Shi Z-L, Fang K, Li Z-H, Ren D-H, Zhang J-Y, et al. EZH2 Inhibition Ameliorates Transverse Aortic Constriction-Induced Pulmonary Arterial Hypertension in Mice (2018): e9174926.
- Kaija H, Pakanen L, Porvari K. RNU6B, a frequent reference in miRNA expression studies, differentiates between deaths caused by hypothermia and chronic cardiac ischemia. Int J Legal Med 134 (2020): 159-162.
- Liu J. Hepatic CREBZF as a novel transcriptional regulator of the lipogenic pathway. Metab Open 1 (2019): 7-8.
- Zhang F, Hu Z, Li G, Huo S, Ma F, et al. Hepatic CREBZF couples insulin to lipogenesis by inhibiting insig activity and contributes to hepatic steatosis in diet-induced insulin-resistant mice. Hepatology 68 (2018): 1361-1375.
- Hu Z, Han Y, Liu Y, Zhao Z, Ma F, et al. CREBZF as a Key Regulator of STAT3 Pathway in the Control of Liver Regeneration in Mice. Hepatology 71 (2020): 1421-1436.
- Lin J, Yoon C, Li P, Ryeom SW, Cho S-J, et al. CDK5RAP3 as tumour suppressor negatively regulates self-renewal and invasion and is regulated by ERK1/2 signalling in human gastric cancer. Br J Cancer (2020): 1-14.
- Li J, Hu X, Su M, Shen H, Qiu W, et al. CDK5RAP3 Participates in Autophagy Regulation and Is Downregulated in Renal Cancer (2019).
- Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE Genes for the Diagnosis of Lung Adenocarcinoma. Int J Biol Markers 22 (2007): 108-113.
- Sixt SU, Peters J. Extracellular Alveolar Proteasome. Proc Am Thorac Soc 7 (2010): 91-96.
- Goggolidou P, Stevens JL, Agueci F, Keynton J, Wheway G, et al. ATMIN is a transcriptional regulator of both lung morphogenesis and ciliogenesis. Development 141 (2014): 3966-3977.
- Torok JA, Oh P, Castle KD, Reinsvold M, Ma Y, et al. Deletion of Atm in Tumor but not Endothelial Cells Improves Radiation Response in a Primary Mouse Model of Lung Adenocarcinoma. Cancer Res 79 (2019): 773-782.
- Foster H, Ruiz EJ, Moore C, Stamp GWH, Nye EL, et al. ATMIN Is a Tumor Suppressor Gene in Lung Adenocarcinoma. Cancer Res 79 (2019): 5159-5166.
- Cho E-A, Kim E-J, Kwak S-J, Juhnn Y-S. cAMP signaling inhibits radiation-induced ATM phosphorylation leading to the augmentation of apoptosis in human lung cancer cells. Mol Cancer 13 (2014): 36.
- Hou H-H, Cheng S-L, Chung K-P, Kuo MY-P, Yeh C-C, et al. Elastase induces lung epithelial cell autophagy through placental growth factor. Autophagy 10 (2014): 1509-1521.
- Aldave AJ, Yellore VS, Vo RC, Kamal KM, Rayner SA, et al. Exclusion of Positional Candidate Gene Coding Region Mutations in the Common Posterior Polymorphous Corneal Dystrophy 1 Candidate Gene Interval. Cornea 28 (2009): 801-807.
- Guo J, Chen X, Xi R, Chang Y, Zhang X, et al. AEG-1 expression correlates with CD133 and PPP6c levels in human glioma tissues. J Biomed Res 28 (2014): 388-395.
