Carboxypeptidase Cathepsin X Defines a Multifunctional Role of Gamma- Enolase in Cancer
Article Information
Tjasa Vi?in Zlobec1, Anja Pišlar1, Ib Jarle Christensen2, Hans Jorgen Nielsen3, Pika Meško Brguljan4, Janko Kos1,5
1Faculty of Pharmacy, University of Ljubljana, Slovenia
2The Finsen Laboratory, Rigshospitalet, Denmark
3Department of Surgical Gastroenterology, Hvidovre Hospital, Hvidovre & Institute of Clinical Medicine, University of Copenhagen, Denmark
4University Clinic for Respiratory and Allergic Diseases Golnik, Slovenia
5Departmnent of Biotechnology, Jošef Stefan Institute, Ljubljana, Slovenia
*Corresponding Author: Janko Kos, Faculty of Pharmacy, University of Ljubljana, Ašker?eva, 1000 Ljubljana, Slovenia.
Received: 27 January 2022; Accepted: 07 February 2022; Published: 28 February 2022
Citation:
Tjaša Vi┼żin Zlobec, Anja Pišlar, Ib Jarle Christensen, Hans Jorgen Nielsen, Pika Meško Brguljan, Janko Kos. Carboxypeptidase Cathepsin X Defines a Multifunctional Role of Gamma- Enolase In Cancer. Journal of Biotechnology and Biomedicine 5 (2022): 20-41.
Share at FacebookAbstract
Gamma-enolase enzymatic activity is involved in glycolysis, a prevalent process in cancer cell metabolism. Additionally, gamma-enolase has a pro-survival function, exhibited through the active site at the C-terminal end of the molecule. This activity is regulated by cysteine peptidase cathepsin X, which cleaves two amino acids at C-terminal end of gamma-enolase. In clinical practice, the determination of gamma-enolase as a tumour marker does not differ between total, uncleaved and C-terminally cleaved forms. However, levels of uncleaved gamma-enolase alone may provide additional clinical information. In this study we analysed cathepsin X, C- terminally uncleaved and total gamma-enolase in tumour cell lines and sera from 255 patients with colorectal cancer (CRC) by western blot, immunoprecipitation, enzymatic activity, ELISAs and ECLIA. Results show that uncleaved gamma-enolase, rather than total gamma- enolase, exhibits different levels in cells, being the highest in those, derived from metastatic sites or highly invasive tumours. Gamma-enolase is secreted into the extracellular space predominantly as an uncleaved form and levels were congruent to those within the cells. Furthermore, levels of uncleaved gamma-enolase in cells are inversely related to cathepsin X protein level and its enzymatic activity. Uncleaved gamma-enolase is also predominant form in sera of patients with CRC. Both forms exhibit similar stage dependent distribution, with slightly elevated levels in stage IV patients. Higher levels of total gamma-enolase are significantly related to shorter survival in patients with metastatic CRC. Results support evidence of additional pro-survival function of gamma-enolase in cancer. Future studies should focus on analysis of uncleaved gamma-enolase in tumour samples, which may provide additional relations to clinical indicators of disease progression.
Keywords
Cancer; Cathepsin X; Cell survival; Gamma-enolase; Prognosis
Cathepsin X articles Cathepsin X Research articles Cathepsin X review articles Cathepsin X PubMed articles Cathepsin X PubMed Central articles Cathepsin X 2023 articles Cathepsin X 2024 articles Cathepsin X Scopus articles Cathepsin X impact factor journals Cathepsin X Scopus journals Cathepsin X PubMed journals Cathepsin X medical journals Cathepsin X free journals Cathepsin X best journals Cathepsin X top journals Cathepsin X free medical journals Cathepsin X famous journals Cathepsin X Google Scholar indexed journals Cell survival articles Cell survival Research articles Cell survival review articles Cell survival PubMed articles Cell survival PubMed Central articles Cell survival 2023 articles Cell survival 2024 articles Cell survival Scopus articles Cell survival impact factor journals Cell survival Scopus journals Cell survival PubMed journals Cell survival medical journals Cell survival free journals Cell survival best journals Cell survival top journals Cell survival free medical journals Cell survival famous journals Cell survival Google Scholar indexed journals Gamma-enolase articles Gamma-enolase Research articles Gamma-enolase review articles Gamma-enolase PubMed articles Gamma-enolase PubMed Central articles Gamma-enolase 2023 articles Gamma-enolase 2024 articles Gamma-enolase Scopus articles Gamma-enolase impact factor journals Gamma-enolase Scopus journals Gamma-enolase PubMed journals Gamma-enolase medical journals Gamma-enolase free journals Gamma-enolase best journals Gamma-enolase top journals Gamma-enolase free medical journals Gamma-enolase famous journals Gamma-enolase Google Scholar indexed journals Prognosis articles Prognosis Research articles Prognosis review articles Prognosis PubMed articles Prognosis PubMed Central articles Prognosis 2023 articles Prognosis 2024 articles Prognosis Scopus articles Prognosis impact factor journals Prognosis Scopus journals Prognosis PubMed journals Prognosis medical journals Prognosis free journals Prognosis best journals Prognosis top journals Prognosis free medical journals Prognosis famous journals Prognosis Google Scholar indexed journals Cancer articles Cancer Research articles Cancer review articles Cancer PubMed articles Cancer PubMed Central articles Cancer 2023 articles Cancer 2024 articles Cancer Scopus articles Cancer impact factor journals Cancer Scopus journals Cancer PubMed journals Cancer medical journals Cancer free journals Cancer best journals Cancer top journals Cancer free medical journals Cancer famous journals Cancer Google Scholar indexed journals enzyme articles enzyme Research articles enzyme review articles enzyme PubMed articles enzyme PubMed Central articles enzyme 2023 articles enzyme 2024 articles enzyme Scopus articles enzyme impact factor journals enzyme Scopus journals enzyme PubMed journals enzyme medical journals enzyme free journals enzyme best journals enzyme top journals enzyme free medical journals enzyme famous journals enzyme Google Scholar indexed journals phosphoenolpyruvate articles phosphoenolpyruvate Research articles phosphoenolpyruvate review articles phosphoenolpyruvate PubMed articles phosphoenolpyruvate PubMed Central articles phosphoenolpyruvate 2023 articles phosphoenolpyruvate 2024 articles phosphoenolpyruvate Scopus articles phosphoenolpyruvate impact factor journals phosphoenolpyruvate Scopus journals phosphoenolpyruvate PubMed journals phosphoenolpyruvate medical journals phosphoenolpyruvate free journals phosphoenolpyruvate best journals phosphoenolpyruvate top journals phosphoenolpyruvate free medical journals phosphoenolpyruvate famous journals phosphoenolpyruvate Google Scholar indexed journals cancer research articles cancer research Research articles cancer research review articles cancer research PubMed articles cancer research PubMed Central articles cancer research 2023 articles cancer research 2024 articles cancer research Scopus articles cancer research impact factor journals cancer research Scopus journals cancer research PubMed journals cancer research medical journals cancer research free journals cancer research best journals cancer research top journals cancer research free medical journals cancer research famous journals cancer research Google Scholar indexed journals gamma-enolase articles gamma-enolase Research articles gamma-enolase review articles gamma-enolase PubMed articles gamma-enolase PubMed Central articles gamma-enolase 2023 articles gamma-enolase 2024 articles gamma-enolase Scopus articles gamma-enolase impact factor journals gamma-enolase Scopus journals gamma-enolase PubMed journals gamma-enolase medical journals gamma-enolase free journals gamma-enolase best journals gamma-enolase top journals gamma-enolase free medical journals gamma-enolase famous journals gamma-enolase Google Scholar indexed journals biologicalá articles biologicalá Research articles biologicalá review articles biologicalá PubMed articles biologicalá PubMed Central articles biologicalá 2023 articles biologicalá 2024 articles biologicalá Scopus articles biologicalá impact factor journals biologicalá Scopus journals biologicalá PubMed journals biologicalá medical journals biologicalá free journals biologicalá best journals biologicalá top journals biologicalá free medical journals biologicalá famous journals biologicalá Google Scholar indexed journals
Article Details
1. Introduction
Enolase is a dimeric intracellular enzyme, responsible for the conversion of 2-phosphoglycerate to phosphoenolpyruvate in the glycolytic pathway. In mammals, there are three isoforms; alpha-, beta-, and gamma-enolase, all expressed in a development-specific manner. Gamma-enolase, known also as Neuron-Specific Enolase (NSE), including two isoenzymes, γγ and αγ, is found predominantly in neurons and neuroendocrine cells [1, 2]. Its increased expression and secretion into the extracellular space is typical for neuroendocrine tumours and has therefore been used as a tumour marker for differential diagnosis, prognosis, therapy monitoring and detection of recurrence in patients with cancers of neuroendocrine origin and others [3-9]. Despite being long known and used as a tumour marker, the role of gamma-enolase in development and progression of cancer is not clear [10, 11]. As a glycolytic enzyme it is involved in increased aerobic glycolysis, the main source of energy in cancer cells [12]. For instance, in malignant transformation of breast [13], urothelial [5] and astrocytic cells [14] increased expression of gamma-enolase was suggested as a critical step in the adaptation to higher metabolic needs of cancer cells [15]. An up-regulation of gamma-enolase expression was detected also in glioblastoma cells, which were cultured under stress conditions, such as serum starvation and hypoxia.
Besides the involvement in glycolysis, a pro-survival function of gamma-enolase has been proposed [16]. The pro-survival function of gamma-enolase has already been demonstrated for neuronal and glial cells [16-19]. It is manifested through the additional active site, which is localized at the C-terminal end of the molecule and is involved in activation of signalling pathways promoting neuronal survival and neuritogenesis [20, 21]. Gamma-enolase pro- survival activity is regulated by cysteine peptidase cathepsin X (Cat X), which is frequently expressed in neuronal and glial cells [22]. As shown by our group, Cat X sequentially cleaves the last two amino acids Leu (433) and Val (432) from the C-terminal end of gamma enolase [23] preventing its binding to gamma-1-syntrophin, a scaffold protein, which mediates trafficking of gamma-enolase towards the plasma membrane [24]. Gamma-enolase association with the plasma membrane proteins was reported to be a prerequisite for its neurotrophic activity [20, 24]. Recently, Trk receptor has been recognized as a binding partner of gamma-enolase in mediating its neurotrophic signalling [25]. The role of tumour marker in addition to gamma enolase is also attributed to Cat X. It was reported as a tumour-promoting factor in several cancer types, being overexpressed in tumour cells and in tumour infiltrated immune cells. It is involved in migration, adhesion, invasion, epithelial-mesenchymal transition and senescence bypass of tumour cells, therefore, supporting development, growth and progression of tumours [26, 27]. This was confirmed in clinical studies, providing relation of higher Cat X mRNA and protein levels to shorter survival of patients with local resectable disease [28-30]. On the other hand, lower Cat X protein levels were related to advanced stage and shorter patient survival [31]. Currently, gamma-enolase, analysed as tumour marker or used in cancer research is not distinguished as intact or C-terminally truncated molecule. C-terminally uncleaved gamma- enolase may act as a pro-survival factor in cancer cells exhibiting significantly different properties as its cleaved form. The aim of the present study was to explore the expression pattern and secretion of uncleaved and total gamma-enolase in different cancer cell lines and to establish their relation to Cat X expression and its activity. Further, we determined the levels of uncleaved gamma-enolase and total gamma-enolase in serum from CRC patients and related them to clinical and pathological parameters.
2. Results
2.1. Specific detection of C-terminally uncleaved gamma-enolase by ELISA
The mouse monoclonal antibody against gamma-enolase, used in our study, recognizes 30- aminoacid peptide, mimicking the uncleaved C-terminal part of the molecule, but not the two- residue-shorter peptide, cleaved by Cat X (Figure 1). Therefore, it can be applied to distinguish between C-terminally uncleaved and cleaved gamma-enolase in tumour cells and used in ELISA to determine uncleaved gamma-enolase in sera from cancer patients.
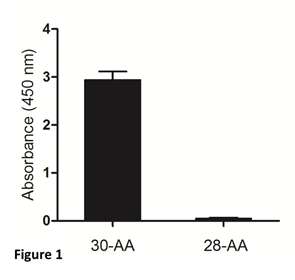
Figure 1: Specificity of the mouse monoclonal antibody for detection of the C-terminally uncleaved gamma-enolase. Two synthetic peptides corresponding to the C-terminal end of human brain gamma-enolase were used. The 30-aminoacid (30-AA) peptide represents the C-terminally uncleaved gamma-enolase and the 28-aminoacid (28-AA) peptide represents the C-terminally cleaved gamma-enolase. The mouse monoclonal antibody against the C-terminally uncleaved gamma-enolase specifically recognizes the full-length protein.
2.2. Expression and secretion levels of uncleaved gamma-enolase differ from the levels of total gamma-enolase and are related to Cat X expression
The ratio between protein levels of uncleaved and total gamma-enolase (U/T ratio) and the relation to Cat X protein level and enzymatic activity was evaluated in cell lysates and supernatants of seven cancer cell lines, namely U87, U373, U251, SH-SY5Y, Caco-2, MCF10A and MCF7. As shown by western blot analysis, uncleaved gamma-enolase exhibits different expression levels in cancer cell lines lysates, while total gamma-enolase is uniformly expressed in all analysed cell lines (Figure 2 A). U/T ratio is the highest in glioblastoma cell line U87 and neuroblastoma cell line SH-SY5Y, which are derived from highly invasive stage IV tumour (U87) [32, 33] or metastatic sites (SH-SY5Y) [34]. On the other hand, a lower U/T ratio can be observed in U373 and U251 glioblastoma astrocytoma cell lines, which are derived from a less invasive (grade III) tumour, compared to U87 cells [33]. Further, colorectal adenocarcinoma cells Caco-2 and non- malignant breast epithelial MCF 10A cells [35] show the lowest U/T ratio, the latter having almost no uncleaved form. MCF7 breast cancer adenocarcinoma cells, which are also derived from a metastatic site [36], show a high expression of uncleaved gamma-enolase, comparable to grade III glioblastoma astrocytoma cell lines levels. To confirm the results obtained from western blot analysis, we measured the levels of uncleaved and total gamma-enolase in cell lysates using direct ELISA (Figure 2B). The U/T ratio of protein levels shows a similar expression pattern as in western blot analysis. Since Cat X processes the C-terminal end of gamma-enolase [23], higher expression of active Cat X in cells could lead to lower levels of uncleaved gamma-enolase. Western blot analysis showed that the levels of active Cat X were indeed inversely related to those of uncleaved gamma-enolase (Figure 2A). Protein levels of total Cat X (pro-form and active form) in cell lysates were evaluated also by ELISA (Figure 2C) and Cat X activity by enzyme kinetics (Figure 2D). Both levels are in accordance with western blot analysis and are inversely related to the uncleaved gamma-enolase levels. Gamma-enolase secretion was further examined, where western blot analysis and ELISA provided a similar protein pattern of uncleaved gamma-enolase in supernatants as in cell lysates (Figure 3 A, B). The same holds for Cat X, with the highest protein levels present in Caco-2 and MCF 10A cell line (Figure 3A, B). Our results confirm that Cat X is secreted from immune cells and cancer cells predominantly as a pro-form (pro-Cat X) [26-29] - as determined by western blot analysis (Figure 3A). Taken together, these data show that uncleaved gamma-enolase and total gamma-enolase have different expression and secretion profiles in different cancer cell lines and that the levels of uncleaved gamma-enolase, but not total gamma-enolase, inversely correlate to the levels of active Cat X in cell lysates. Higher levels of Cat X may therefore be related with more intensive gamma-enolase C-terminal end processing. Uncleaved gamma-enolase seems to be the predominant form to be secreted from cells and its levels reflect well those found in cell extracts. Extracellular forms are not dependent on Cat X, which is present in supernatants predominantly as inactive pro-enzyme.
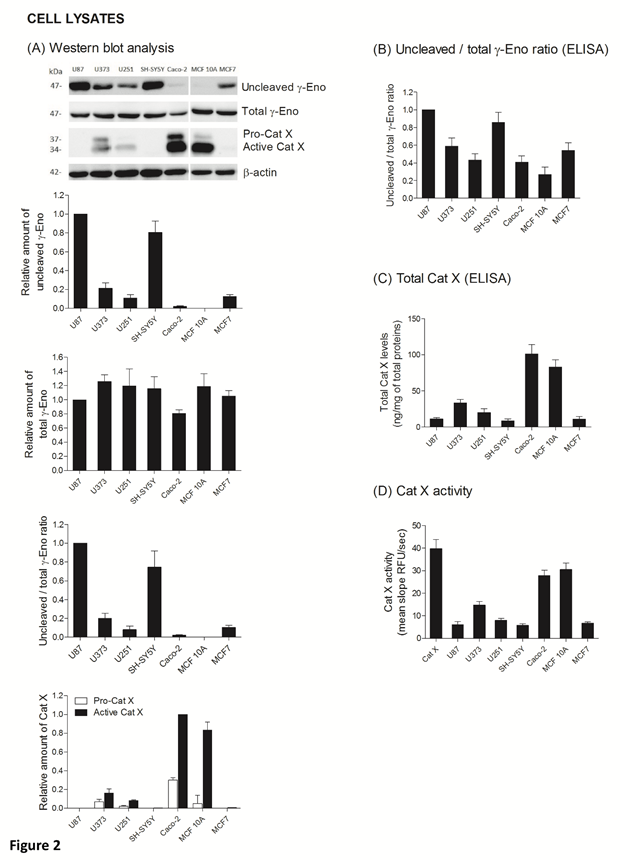
Figure 2: Uncleaved and total gamma-enolase have different expression profiles in cell lines and are related to Cat X expression. Western blot analysis of uncleaved and total gamma-enolase and Cat X in U87, U373, U251, SH-SY5Y, Caco-2, MCF 10A and MCF7 cell lysates. While total gamma- enolase is uniformly expressed in all analysed cell lines, uncleaved gamma-enolase has different expression levels. Cat X is expressed mainly as active enzyme and its expression is inversely related to uncleaved gamma-enolase expression: the highest expression of active Cat X can be detected in cell lines with the lowest uncleaved gamma-enolase expression. Graphs below western blot images indicate the relative protein amount of uncleaved gamma-enolase, total gamma-enolase, uncleaved/total gamma enolase ratio (U/T ratio) and pro- or active Cat X. (A). Using ELISA, we measured the values of uncleaved gamma-enolase, total gamma-enolase and total Cat X in cell lysates to confirm the results from western blotting. The U/T protein amount ratio (B) and levels of total Cat X (C) are in accordance with the results from western blot analysis. Total Cat X values are presented as ng of total Cat X per mg of total protein amount in the cell lysate. Cat X activity was demonstrated using enzyme kinetics. 30 nM human recombinant Cat X was used as a positive control (D). Values are given as means of three independent biological replications ±SD. Abbreviations: γ-Eno; gamma-enolase, Pro-Cat X; pro- cathepsin X, Active Cat X; active cathepsin X.
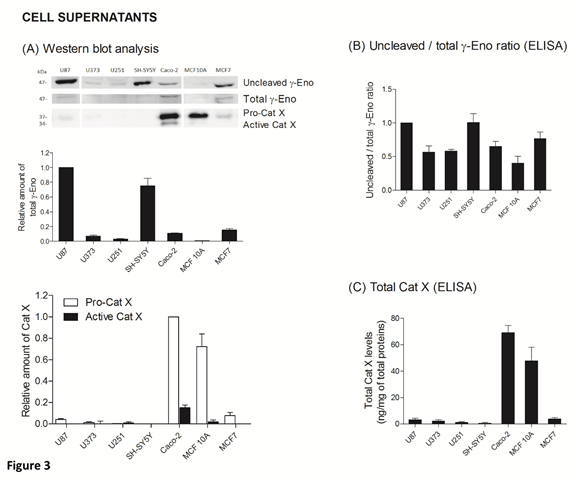
Figure 3: Uncleaved and total gamma-enolase have different expression profiles in cell supernatants. Western blot analysis of uncleaved and total gamma-enolase and Cat X in U87, U373, U251, SH-SY5Y, Caco-2, MCF 10A and MCF7 cell supernatants. Graphs below western blot images indicate the relative protein amount of uncleaved gamma-enolase and pro- or active Cat X (A). Uncleaved gamma-enolase, total gamma-enolase and total Cat X in cell supernatants were measured also by ELISAs. Total Cat X values are presented as ng of total Cat X per mg of total protein amount in cell supernatants. Values are given as means of three independent biological replications ±SD. Abbreviations: γ-Eno; gamma-enolase, Pro-Cat X; pro-cathepsin X, Active Cat X; active cathepsin X.
2.3. Uncleaved and total gamma-enolase have different expression profiles in human sera
To assess the U/T ratio of gamma-enolase in sera from five patients with CRC, we immunoprecipitated total gamma-enolase with goat polyclonal antibody. Immunoprecipitates were further resolved by SDS-PAGE and analysed on western blot: first with mouse monoclonal antibody against uncleaved gamma-enolase and then with mouse monoclonal antibody against total gamma-enolase. As shown in Figure 4, the amounts of uncleaved and total gamma-enolase differ between the samples, however, U/T ratio remains similar. Therefore, the uncleaved gamma-enolase might be a predominant form in patient sera.
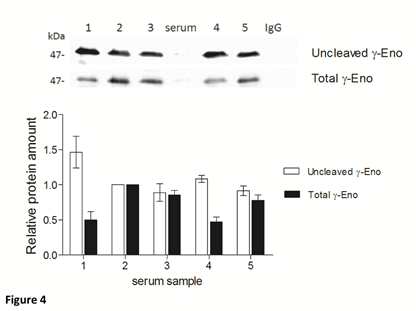
Figure 4: Uncleaved and total gamma-enolase have different expression profiles in human sera. Immunoprecipitation of total gamma-enolase from five sera from patients with colorectal cancer. Immunoblots were resolved with SDS-PAGE and Western blot. The graph below western blot images indicates the relative protein amounts of uncleaved and total gamma-enolase. As a negative control nonspecific goat polyclonal IgG was used. Besides the immunoprecipitates, we applied also a serum sample (denoted »serum«) directly on the gel, however, the values of gamma-enolase were below the detection limit. Data are means of two independent experiments ± SD. Abbreviations: γ-Eno; gamma- enolase.
2.4. Levels of uncleaved gamma-enolase, total gamma-enolase, and Cat X in sera of patients with CRC and association with baseline variables
Protein levels of uncleaved gamma-enolase, total gamma-enolase and Cat X were measured in sera from 264 patients with CRC. Statistical analysis was performed for 255 samples, due to haemolysis detected in nine samples. Values and descriptive statistics are shown in Table 2. According to Roche Diagnostics GmbH Elecsys NSE assay datasheet, the expected value of total gamma-enolase in healthy subjects is lower than 16.3 ng/ml (95% Confidence Interval (CI): 15.7-17.0 ng/ml). In our study, the mean value of total gamma-enolase in CRC patients was 14.3 ng/ml, while the mean value of uncleaved gamma-enolase was 10.3 ng/ml. Uncleaved and total gamma-enolase values were stratified by stage. They show similar, stage dependent distribution with slightly elevated levels in stage IV patients (Figure 5), however, the difference is statistically significant for total (p<0.0001) but not for uncleaved gamma- enolase (p>0.09).
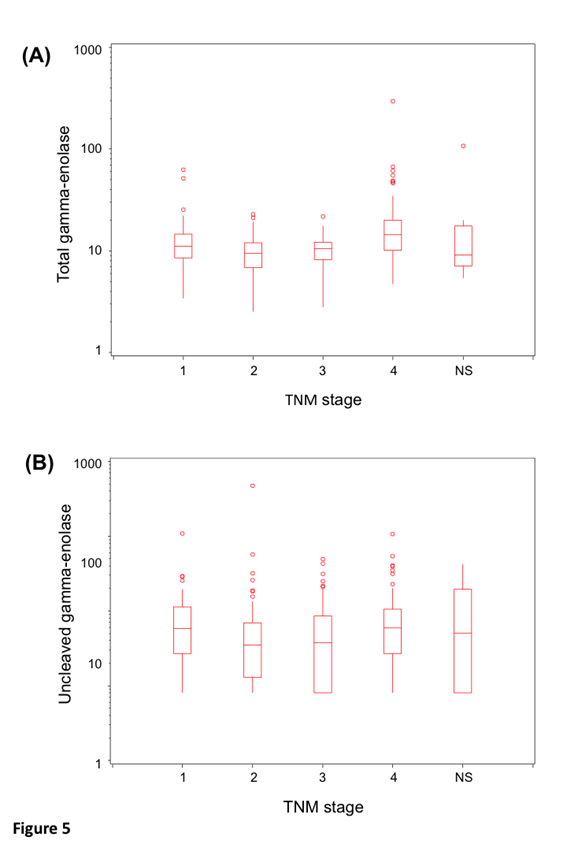
Figure 5: Distribution of uncleaved and total gamma-enolase values by stage. Box plots show the lower and upper quartiles and the median. The whiskers are maximal 1.5 times the interquartile range. The distribution of uncleaved gamma-enolase (A) and total gamma-enolase (B) values by stage. NS denotes patients, which have not been staged.
2.5. Analysis of survival
Univariate analysis of uncleaved and total gamma-enolase for the entire set showed that higher levels of both, uncleaved (HR=1.10; 95% CI: 1.01-1.21; p=0.0363) and total gamma-enolase (HR=1.86; 95% CI: 1.46-2.37; p<0.0001) are significantly related to shorter survival of patients with CRC. Multivariate analysis of the entire set included as covariates total gamma-enolase, age (stratified per 10 years), gender, stage, and tumour localisation (Table 3). A significant interaction was demonstrated between total gamma-enolase and stage (p=0.038). Total gamma-enolase was associated to outcome in patients in stage IV only (HR: 1.77, 95% CI: 1.40-2.24; p<0.0001). Further, multivariate analysis was performed separately for patients at stages I-III and those with stage IV, as previously reported [29]. Analysis of patients in stages I-III could not demonstrate a significant association of total gamma-enolase to outcome (HR=0.90, 95% CI: 0.58-1.39, p=0.64), however analysis of stage IV patients showed that total gamma- enolase was significantly associated to overall survival (HR=2.02 95% CI: 1.49-2.75, p<0.0001). Treatment with chemotherapy has not been included in the analysis for stages I-III, since primarily only patients in stage III received treatment (p=0.97). On the other hand, analysis for stage IV included treatment with chemotherapy, but no interaction between total gamma-enolase and chemotherapy could be demonstrated (p=0.48). It has to be emphasized that these patients were not randomized to palliative chemotherapy, but selected as part of the clinical decisions. Kaplan-Meier curves provide a similar result for patients in stage IV using tertiles as the cut off value (Figure 6). Taken together, in sera from patients with CRC, total gamma-enolase has a prognostic value for metastatic CRC.
|
Gender |
Subjects, n (%) |
|
|
Female |
104 (40.8) |
|
|
Male |
151 (59.2) |
|
|
Age (years) |
70.3, 32.7 - 93.3 (median, range) |
255 (100.0) |
|
Localisation |
||
|
Left-sided colon cancer |
96 (37.7) |
|
|
Right-sided colon cancer |
58 (22.8) |
|
|
Rectal cancer |
101 (39.6) |
|
|
Stage |
I |
42 (16.5) |
|
II |
70 (27.5) |
|
|
III |
62 (24.3) |
|
|
IV |
66 (25.9) |
|
|
Not staged |
15 (5.9) |
|
|
Administered chemotherapy |
5-FU |
1 (0.4) |
|
5-FU + Isovorin |
17 (6.7) |
|
|
Irinotecan |
1 (0.4) |
|
|
Irinotecan + 5-FU Irinotecan + 5-FU + Calcium folinat + Avastin |
7 (2.8) |
|
|
3 (1.2) |
||
|
Oxaliplatin + 5-FU Oxaliplatin + 5-FU + Cetuximab |
11 (4.3) |
|
|
3 (1.2) |
||
|
Capecitabine |
6 (2.4) |
|
|
Oxaliplatin + Capecitabine |
27 (10.6) |
|
|
Other |
4 (1.6) |
|
|
Total |
80 (31.4) |
Table 1: Patient characteristics and applied first line therapies.
|
Variable |
Median |
Minimum |
Maximum |
|
uncleaved gamma-enolase |
4.54 |
0.8 |
469.26 |
|
total gamma-enolase |
10.8 |
2.5 |
295.3 |
|
total Cat X |
23.18 |
8.69 |
47.14 |
|
Values are given as ng/ml. |
|||
Table 2: Descriptive statistics.
|
Parameter |
Stages I-III |
HR |
95% CI |
p-value |
|
Gamma-enolase |
2-fold difference in HR |
0.9 |
0.58-1.39 |
0.6397 |
|
Age (per 10 years) |
Per 10 years |
1.52 |
1.19-1.93 |
0.0007 |
|
Gender |
F vs. M |
0.7 |
0.41-1.19 |
0.1849 |
|
Localisation |
Left-sided colon cancer |
1.13 |
0.59-2.18 |
0.7174 |
|
Rectal cancer |
0.84 |
0.42-1.68 |
0.6161 |
|
|
Stage |
II vs. I |
1.1 |
0.51-2.35 |
0.8152 |
|
III vs. I |
2.38 |
1.18-4.79 |
0.0152 |
|
|
Stage IV |
||||
|
Gamma-enolase |
2-fold difference in HR |
2.02 |
1.49-2.75 |
<0.0001 |
|
Chemotherapy |
Yes vs. No |
0.17 |
0.08-0.34 |
<0.0001 |
|
Age (per 10 years) |
Per 10 years |
1.01 |
0.72-1.40 |
0.965 |
|
Gender |
F vs. M |
0.67 |
0.39-1.16 |
0.1531 |
|
Localisation |
Left-sided colon cancer |
0.23 |
0.11-0.48 |
<0.0001 |
|
Rectal cancer |
0.27 |
0.12-0.61 |
0.0016 |
Table 3: Multivariate analysis for overall survival of patients in stages I-III and IV.
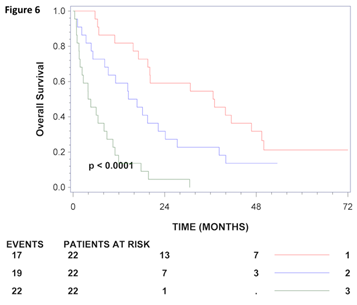
Figure 6: Kaplan-Meier survival curves for CRC patients in stage IV. CRC patients in stage IV were grouped according to gamma-enolase values using tertiles as tresholds. The number of patients at risk at 0, 24 and 48 months for each group is shown below the axis for each stratum and the number of deaths (events) is shown to the left. Kaplan-Meier estimates of survival for patients in stage IV who, 1: had total gamma-enolase levels in the lowest tertile, 2: had total-gamma enolase levels in the middle tertile 3: had total gamma-enolase levels in the highest tertile.
3. Discussion
We evaluated protein levels of C-terminally uncleaved gamma-enolase, which possesses the pro-survival active site, in various cell lines and compared it to the levels of total gamma- enolase. The results show that uncleaved gamma-enolase rather than total gamma-enolase, exhibits different expression profiles, and that gamma-enolase is secreted into the extracellular space predominantly as an uncleaved form. Furthermore, the levels of uncleaved gamma- enolase inversely correlated to the active form of Cat X, confirming gamma-enolase as a possible target for Cat X activity.
The levels of uncleaved gamma-enolase in sera of patients with CRC were similar to those of total gamma-enolase, due to secretion of uncleaved form, as observed on cell lines and the fact that also Cat X is secreted as an inactive pro-form. The most known and studied tumour supportive function of enolase is the involvement in enhanced glycolysis in cancer cells [37]. When tumours outgrow their blood supply, tumour cells have to adapt to a niche, dominated by hypoxia and lack of nutrients. The lack of oxygen causes a metabolic switch to energetically less efficient processing of pyruvate to lactate, however cancer cells favour this mechanism, also called the “Warburg effect”, regardless the presence of oxygen [38, 39]. Enhanced glycolysis including higher enolase activity is a hallmark of this cancer associated metabolic switch. Several enzymes of the glycolytic pathway, including enolase, are capable of functioning in different physiological and pathological processes possessing besides enzymatic role also other functions [10]. Alpha- enolase, for example, was demonstrated to interact with a variety of nuclear, cytoplasmic or membrane molecules and therefore play a pleiotropic role in cancer progression [37, 40]. The multifunctional role of gamma-enolase, especially in processes of malignant progression, has been explored far less [11]. The first evidence suggesting its multifunctional role in cancer was a significant difference observed between gamma-enolase enzyme activity and protein levels in small-cell lung cancer cell lines. Authors proposed that enzymatically inactive compound may possess other functions, such as cell growth and differentiation [41]. Additionally, nuclear localisation of generally cytoplasmic gamma-enolase in various cancer cells suggested functions other than participation in the glycolytic pathway [13, 42]. In neuronal cells, C-terminally uncleaved gamma-enolase was shown to promote their survival, differentiation, regeneration and to impair apoptosis by activating phosphatidylinositol 3-kinase (PI3K) and Mitogen-Activated Protein Kinase (MAPK) signal transduction pathways [20] or by suppressing the activation of p75 neurotrophin receptor (p75NTR) downstream effectors in apoptotic signalling [43]. This effect is regulated by Cat X, which cleaves the gamma-enolase active site, responsible for the pro-survival function [23, 24].
In cancer cells, the mechanism of the pro-survival function of gamma-enolase has not been explored so far. In general, increased levels of gamma-enolase were shown to be related to cancer progression and being typical for advanced stages and for distant metastases [44, 45]. In malignantly transformed urothelial cells, higher gamma-enolase expression was associated to cell proliferation and colony formation [5]. Exposure of glioblastoma cell lines to hypoxia and serum starvation caused increased gamma-enolase mRNA or protein levels and, vice versa, gamma-enolase knock-down sensitized cells to hypoxia, radiotherapy and chemotherapy. In all these studies total gamma-enolase was presumably detected, including uncleaved and cleaved forms. Uncleaved gamma-enolase might have stronger clinical utility, since our results show higher concentrations of uncleaved gamma-enolase in cells, derived from metastatic sites or from invasive tumours. Tumour cells that migrate from primary tumours to metastatic sites may utilize uncleaved gamma-enolase to survive a niche with lack of oxygen and nutrients and later to proliferate and forming a secondary tumour. Expression of active Cat X in cancer cell lines is inversely related to uncleaved gamma-enolase and is the lowest in cells, derived from metastatic sites or invasive tumours. These results are in accordance to a study by Jechorek et al., which showed that Cat X expression is the lowest in metastatic disease and that the loss of Cat X is essential for tumour cell detachment, local invasion and tumour progression [31]. In the advanced stages of cancer, loss of Cat X in cells might therefore have a dual role: to enable cell detachment and invasion and to keep higher rates of uncleaved gamma-enolase in tumour cells promoting their survival at unfavourable conditions. Our study further confirms previous results that Cat X is secreted from cells into supernatants mainly as a pro-enzyme and cannot process extracellular intact gamma- enolase [26, 29]. We found the highest secretion of Cat X from Caco-2 and MCF 10A cells, which, among all tested cells, already showed the highest intracellular concentrations. Therefore, extracellular levels of Cat X may reflect the expression within the cells and most probably to a lower extent the differences in secretion between the cells. However, it has to be stressed, that within tumour microenvironment, Cat X is secreted also from tumour associated immune cells and that the effects of Cat X could be as well a consequence of tumour-immune cells interactions [27, 31, 46]. Currently, gamma-enolase, used as a serum-based marker for monitoring the outcome of treatment and disease course in cancer patients [11], is not distinguished as intact and C-terminally truncated molecule. We propose that the truncation of gamma-enolase by Cat X could be an important event inside tumour cells with a significant impact on survival and proliferation, whereas extracellular gamma-enolase is predominantly uncleaved. In CRC, moderately elevated serum levels of gamma-enolase have been reported and related to pathological tumour-node-metastasis staging, lymph node metastasis and distant metastasis [47]). Higher gene expression levels have also been related to shorter survival [48]. In our study the levels of uncleaved and total gamma-enolase in patient’s sera were not significantly elevated compared to reference values of healthy controls. However, higher levels of gamma-enolase correlated with shorter survival in patients with metastatic CRC. Uncleaved gamma-enolase revealed similar relations to clinical parameters as the total gamma-enolase, suggesting that in sera total gamma-enolase practically consists of uncleaved gamma-enolase. On the same cohort of patients, higher Cat X levels were related to shorter survival only in patients with local resectable disease, but not in those who had metastatic CRC [29], which might be due to a loss of Cat X in order to support the invasion of cells and the function of gamma-enolase.
Our study provides a new insight into the widely used tumour marker gamma-enolase. The C- terminally uncleaved gamma-enolase, which possesses an additional, pro-survival function, exhibits different expression levels in tumour cells, compared to total gamma-enolase, and is inversely related to Cat X expression. Uncleaved gamma-enolase is a predominant form in cell supernatants as well as in sera from patients with CRC. Only in a group of patients with metastatic CRC serum gamma-enolase correlated with survival. Further studies should focus on the analysis of uncleaved gamma-enolase in tumour samples, which may provide additional relations to clinical indicators of disease progression and enable the selection of patients with more aggressive tumor phenotype.
4. Materials and Methods
4.1. Cell culture
Human glioblastoma (U-87 MG), human glioblastoma astrocytoma (U-373 MG and U-251 MG), human neuroblastoma (SH-SY5Y), human colorectal adenocarcinoma (Caco-2), human breast cancer adenocarcinoma (MCF7) and human non-malignant breast epithelial cells (MCF 10A) were purchased from American Type Culture Collection (ATCC, Manassas, VA, U.S.). U-87 MG, U-373 MG, U-251 MG and Caco-2 cells were grown in Eagle's Minimum Essential Medium (Sigma-Aldrich, St. Louis, MO, U.S.), supplemented with 10 % (v/v) fetal bovine serum (HyClone, GE Healthcare, Chicago, IL, U.S.). MCF7 and MCF 10A cells were grown in Eagle's Minimum Essential Medium (Sigma-Aldrich), supplemented with 10 % (v/v) fetal bovine serum (HyClone), 10 μg/ml insulin, 0.5 μg/ml hydrocortisone and 20 ng/ml EGF (all from Sigma-Adrich). SH-SY5Y cells were grown in Dulbecco`s modified Eagle`s medium (Sigma-Aldrich), supplemented with 10 % (v/v) fetal bovine serum. All described cells were supplemented with 1 % L-glutamine and 1 % penicillin/streptomycin (all from Sigma-Adrich) and maintained at 37 °C in a humidified atmosphere with 5% CO2.
4.2. Patients and sampling
The multi-centre observational cohort study was conducted at six Danish hospitals from October 2003 through December 2005 and included 264 subjects diagnosed with primary rectal or colon cancer. All subjects gave informed consent for inclusion into the study. The median time from inclusion to the end of study was 4.8 years (3.9-6.0). Clinical study characteristics and sampling were described previously [29]. Nine samples were excluded from statistical analysis due to haemolysis. Patient characteristics and applied chemotherapy are given in (Table 1). The study was approved by Regional Ethical Committee of Copenhagen (H- 3-2009-110) and The Danish Data Protection Agency (2007-58-0015).
4.3. Antibody specificity analysis
The specificity of the mouse monoclonal antibody against C-terminally uncleaved gamma- enolase (Santa Cruz Biotechnology, Dallas, TX, U.S.), was determined by “in house” indirect ELISA with two synthetic peptides corresponding to the C-terminal end of human brain gamma-enolase, which were synthesized by Biosynthesis (Lewisville, TX, U.S.) and described and used previously [20]. The 30-amino acid peptide represented the C- terminally uncleaved (active) gamma-enolase and a two-amino acid shorter peptide represented the C-terminally cleaved (inactive) gamma-enolase. Buffers used were described previously [28, 29]. Microtiter plates (Nunc-IMMUNO™ MODULES) were coated with 5 μg/mL of each peptide and incubated over night at 4oC. Plates were washed and blocked with 2% BSA for 1h. Then, mouse monoclonal antibody against the C-terminal end of gamma-enolase was added and incubated for 2h at 37oC. After washing, HRP-conjugated anti- mouse secondary antibody (1:3000; Merck Millipore, Burlington, MA, U.S.) was applied and incubated for additional 1.5h at 37oC. Finally, after washing, TMB (3,3´,5,5´- tetramethylbenzidine) Liquid Substrate System (Sigma-Aldrich) was added and after 15 minutes the reaction was stopped with 2 M H2SO4. The absorbance was measured at 450 nm with a microplate reader Tecan Safire2 (Männedorf, Switzerland) and corrected for the absorbance of control.
4.4. Western blot analysis
For detection of proteins in cell lysates and protein secretion in cell supernatants, cells were grown to reach 80% confluence. Cell supernatants were centrifuged and concentrated (Amicon® Ultra, Merck Millipore). To prepare cell lysates, cells were washed with PBS, pH 7.4, and then lysed with 100 µl lysis buffer (50 mM HEPES pH 6.5, 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100) with protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, U.S.) added and left for 30 minutes on ice. Lysates were sonicated and centrifuged at 14000 g at 4°C for 15 min. Total protein concentration was determined by DC Protein Assay (Bio-Rad Laboratories, Hercules, CA, U.S.). Equal amounts of protein (30 µg per lane for cell lysates and 100 µg per lane for concentrated cell supernatants) were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were then blocked with 5 % (m/v) skimmed milk powder in TBST (Tris-buffered saline with Tween 20: 25 Mm Tris/HCl, 137 mM NaCl, 3 mM KCl, pH 7.4, and 0.1 % Tween 20) for 1 h at room temperature. Membranes were incubated with primary antibody in TBST with 3 % (w/v) BSA overnight at 4°C to detect corresponding proteins. The antibodies used were mouse monoclonal antibody against active uncleaved gamma-enolase (1:500), mouse monoclonal antibody against total gamma-enolase (recognizing a region in the internal part of gamma-enolase) (1:500), goat polyclonal antibody against total gamma-enolase (recognizing a region in the N-terminal part of gamma-enolase) (1:250), β-actin (1:500) (all Santa Cruz Biotechnology) or goat polyclonal antibody against Cat X, which recognize both, Cat X pro-from and active form (1:1000) (R&D Systems, Minneapolis, MN, U.S.). For Cat X detection, 1.5 % (m/v) skimmed milk powder and 1 % BSA in TBST was used to block the membranes and to dilute the primary antibody. Membranes were washed three times for 10 min in TBST and then incubated for 1.5 h with corresponding Horseradish Peroxidase (HRP)-conjugated secondary antibody in TBST with 5 % (m/v) skimmed milk powder. Finally, the membranes were washed three times for 10 min in TBST. The bands were visualized with Super Signal chemiluminiscent substrate (Thermo Fisher Scientific) and G-BOX (Syngene, Frederick, MD, U.S.). Band intensities were quantified using Syngene`s Gene Tools Software.
4.5. Immunoprecipitation of gamma-enolase from serum
To detect the amounts of uncleaved and total gamma-enolase in serum samples we randomly selected five serum samples for immunoprecipitation of total gamma-enolase from serum. To 200 μl of each serum we added 5 μg of goat polyclonal antibody against total gamma-enolase (recognizing a region in the N-terminal part of gamma-enolase; Santa Cruz Biotechnology) and mixed overnight at 4oC. As a negative control, nonspecific goat IgG was used. To purify the immune complexes, 50 μl of protein A-Sepharose beads (Amersham Pharmacia Biotech AB, Staffanstorp, Sweden) were added and mixed for 2 h at 4oC. Then, samples were centrifuged for 1 min at 2000 g and washed with binding buffer (0.14 M phosphate buffer, pH 8.2). Finally, immune complexes were isolated by boiling in SDS sample buffer. Then, samples were resolved by SDS-PAGE and immunoblotted as described above.
4.6. Cathepsin X activity
Cathepsin X activity in cell lysates and cell supernatants was measured with the cathepsin X- specific fluorogenic substrate Abz-Phe-Glu-Lys (Dnp)-OH [49]. Cell lysates were prepared in lysis buffer (0.05 M sodium acetate, pH 5.5, 0.1 M NaCl, 1 mM EDTA, 0.25% Triton X-100) and total protein concentrations were determined by DC Protein Assay (Bio-Rad Laboratories). Samples with 25 μg of proteins (in 100 mM acetate buffer, pH 5.5, 0.1% (w/v) polyethylene glycol 8000, 5 mM DTT and 1.5 mM EDTA), were prepared and added to 5 mM substrate in black microplate wells and incubated at 37oC. The formation of fluorescent degradation products was monitored continuously on a microplate reader (Tecan Safire2) at an excitation wavelength of 320 nm and emission wavelength of 420 nm. The signal was corrected for the fluorescence of control.
4.7. Quantitative analysis of uncleaved and total gamma-enolase and Cat X
To measure uncleaved gamma-enolase, microtiter plates were coated with 2 μg/mL of mouse monoclonal antibody against human uncleaved gamma-enolase and incubated over night at 4oC. Plates were washed and blocked with 2% BSA for 1h. Then, gamma-enolase standards (EMELCA Bioscience, Clinge, The Netherlands), cell lysates (25 μg of proteins), cell supernatants (100 μg of proteins) or serum samples (diluted in a 1:2 ratio with dilution buffer) were applied and incubated for 2h at 37oC. Again, plates were washed and mouse monoclonal HRP-conjugated antibody against gamma-enolase (Abcam, Cambridge, UK) was applied (1:2000) and incubated for 1.5h. Finally, plates were washed and LuminataTM Forte ELISA HRP Substrate (Merck Millipore) was added and the signal was measured according to manufacturer instructions, with a microplate reader (BioTek Synergy H4, Winooski, VT, U.S.). The signal was corrected for the absorbance of control. The buffers, the procedure and validation of the ELISA were described previously [29]. The range of the calibration curve extended from 0.4 to 50 ng/ml (Supplemental information 1). The detection limit of the assay was determined to be 0.4 ng/ml. Dilutions of four serum samples, diluted from 1:2 to 1:16, showed a linear response, parallel to the calibration curve (Supplemental information 2). Recovery levels were determined by comparing the observed and expected concentrations of gamma-enolase. Recovery levels varied from 86.5 to 98.1%. Mean recovery was 92.4%. Results are shown in Supplemental information 3. The intra-assay Coefficient of Variance (CV) was 5.5% and the inter-assay CV was determined to be 5.7%. Total gamma-enolase levels were measured on the same way using a mouse monoclonal antibody to an internal part of human gamma-enolase (Santa Cruz Biotechnology) as a capture antibody. Total gamma-enolase in sera was measured at the University Clinic for Respiratory and Allergic Diseases Golnik, Slovenia, with the electrochemiluminiscence automated immunoassay system (ECLIA) using the Elecsys NSE assay and a Elecsys 2010 immunoassay analyzer (Roche Diagnostics GmbH, Rotkreuz, Switzerland). The measuring range of the assay is from 0.050 to 370 ng/ml. The specific performance data of the assay are described in Roche Diagnostics GmbH Elecsys NSE assay datasheet. Total Cat X was determined by ELISA as described [29]. All determinations in sera from cancer patients were performed blinded to the study endpoint.
5. Statistical Analysis
Distribution of the values of uncleaved and total gamma-enolase by stages was analysed and represented with box plots, which show the lower and upper quartiles and the median. The whiskers are maximal 1.5 times the interquartile range. A linear model was used to test the differences in uncleaved and total gamma-enolase levels between stages. Univariate and multivariate Cox regression analysis was done in order to determine association of uncleaved and total gamma-enolase levels and other clinical and pathological parameters with overall survival of CRC patients. Both, uncleaved and total gamma-enolase, were scored as continuous log transformed covariates (using base 2), meaning that hazard ratios (HR) are for two fold differences in the markers levels. Model assessment was done using cumulated sums of martingale residuals. All results are presented with 95% confidence limits and p-values less than 5% are considered significant. Statistical analysis of the clinical study was done with SAS (v9.3, SAS Institute, Cary, N.C., USA) and R (R Core Team (2013). R: A language and environment for statistical computing, R statistical computing, Vienna, Austria. URL. http.//www.R-project.org/). These results are reported in accordance with the REMARK guidelines [50].
Conflict of interest
All authors have read the editorial's policy and declare that they have no competing interests or conflicts of interest of a financial nature.
Funding
This work was supported by Slovenian Research Agency (grants P4-0127, J4-0123 to J.K.). HJN received financial support from The Danish Cancer Society, The Kornerup Fund, The Aage and Johanne Louis-Hansen Fund, The Aase and Ejnar Danielsen Fund, and The Kathrine and Vigo Skovgaard Fund. The sponsors were not involved in the study design, in the collection, analysis and interpretation of data, in the writing of the report or in the decision to submit the article for publication.
References
- Schmechel D, Marangos P, Brightman M. Neurone-specific enolase is a molecular marker for peripheral and central neuroendocrine cells. Nature 276 (1978): 834-836.
- Schmechel DE, Marangos PJ, Martin BM, et al. Localization of Neuron-Specific Enolase (NSE) mRNA in human brain. Neuroscience Letters 76 (1987): 233-238.
- Fujiwara H, Arima N, Ohtsubo H, et al. Clinical significance of serum neuron-specific enolase in patients with adult T-cell leukemia. American Journal of Hematology 71 (2002): 80-84.
- Li L, Zhang Z, Hu Y. Neuron - specific enolase predicts the prognosis in advanced small cell lung cancer patients treated with first-line PD-1/PD-L1 inhibitors. Medicine 100 (2021): 27029.
- Soh M, Dunlevy JR, Garrett SH, et al. Increased neuron specific enolase expression by urothelial cells exposed to or malignantly transformed by exposure to Cd2+ or As3+. Toxicology Letters 212 (2012): 66-74.
- Sturgeon C. Practice guidelines for tumor marker use in the clinic. Clinical Chemistry 48 (2002): 1151-1159.
- Sturgeon CM, Duffy MJ, Stenman UH, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clinical Chemistry 54 (2008): 11-79.
- Tapia FJ, Barbosa AJA, Marangos PJ, et al. Neuron-specific enolase is produced by neuroendocrine tumours. Lancet (London, England) 1 (1981): 808-811.
- Wang L, Liu P, Chen X, et al. Serum neuron-specific enolase is correlated with clinical outcome of patients with non-germinal center B cell-like subtype of diffuse large B- cell lymphoma treated with rituximab-based immunochemotherapy. Medical Oncology (Northwood, London, England) 29 (2012): 2153-2158.
- Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cellular and Molecular Life Sciences: CMLS 58 (2001): 902-920.
- Vizin T, Kos J. Gamma-enolase: a well-known tumour marker, with a less-known role in cancer. Radiology and Oncology 49 (2015): 217-226.
- Jang SM, Kim JW, Kim CH, et al. p19 (ras) Represses proliferation of non-small cell lung cancer possibly through interaction with Neuron-Specific Enolase (NSE). Cancer Letters 289 (2010): 91-98.
- Soh MA, Garrett SH, Somji S, et al. Arsenic, cadmium and neuron specific enolase (ENO2, γ-enolase) expression in breast cancer. Cancer Cell International 11 (2011): 41.
- Vinores SA, Bonnin JM, Rubinstein LJ, et al. Immunohistochemical demonstration of neuron-specific enolase in neoplasms of the CNS and other tissues. Archives of Pathology & Laboratory Medicine 108 (1984a): 536-540.
- Vinores SA, Bonnin JM, Rubinstein LJ, et al. Immunoradiometric and immunohistochemical demonstration of neuron-specific enolase in experimental rat gliomas. Cancer Research 44 (1984b): 2595-2599.
- Hattori T, Takei N, Mizuno Y, et al. Neurotrophic and neuroprotective effects of neuron-specific enolase on cultured neurons from embryonic rat brain. Neuroscience Research 21 (1995): 191-198.
- Hafner A, Glavan G, Obermajer N, et al. Neuroprotective role of γ-enolase in microglia in a mouse model of Alzheimer’s disease is regulated by cathepsin X. Aging Cell 12 (2013): 604-614.
- Pišlar AH, Kos J. C-terminal peptide of γ-enolase impairs amyloid-β-induced apoptosis through p75NTR signaling. NeuroMolecular Medicine 15 (2013a): 623-635.
- Takei N, Kondo J, Nagaike K, et al. Neuronal Survival Factor from Bovine Brain Is Identical to Neuron-Specific Enolase. Journal of Neurochemistry 57 (1991): 1178-1184.
- Hafner A, Obermajer N, Kos J. γ-Enolase C-terminal peptide promotes cell survival and neurite outgrowth by activation of the PI3K/Akt and MAPK/ERK signalling pathways. The Biochemical Journal 443 (2012): 439-450.
- Hattori T, Ohsawa K, Mizuno Y, et al. Synthetic Peptide Corresponding to 30 Amino Acids of the C-Terminal of Neuron-Specific Enolase Promotes Survival of Neocortical Neurons in Culture. Biochemical and Biophysical Research Communications 202 (1994): 25-30.
- Wendt W, Zhu XR, Lübbert H, et al. Differential expression of cathepsin X in aging and pathological central nervous system of mice. Experimental Neurology 204 (2007): 525-540.
- Obermajer N, Doljak B, Jamnik P, et al. Cathepsin X cleaves the C- terminal dipeptide of alpha- and gamma-enolase and impairs survival and neuritogenesis of neuronal cells. The International Journal of Biochemistry & Cell Biology 41 (2009): 1685-1696.
- Hafner A, Obermajer N, Kos J. Gamma-1-syntrophin mediates trafficking of gamma- enolase towards the plasma membrane and enhances its neurotrophic activity. NeuroSignals 18 (2011): 246-258.
- Pišlar A, Kos J. γ-Enolase enhances Trk endosomal trafficking and promotes neurite outgrowth in differentiated SH-SY5Y cells. Cell Communication and Signaling: CCS 19 (2021): 118.
- Kos J, Vi?in T, Fonovi? UP, et al. Intracellular signaling by cathepsin X: molecular mechanisms and diagnostic and therapeutic opportunities in cancer. Seminars in Cancer Biology 31 (2015) 76-83.
- Akkari L, Gocheva V, Kester JC, et al. Distinct functions of macrophage-derived and cancer cell-derived cathepsin Z combine to promote tumor malignancy via interactions with the extracellular matrix. Genes and Development 28 (2014): 2134-2150.
- Vizin T, Christensen I, Nielsen H, et al. Cathepsin X in serum from patients with colorectal cancer: relation to prognosis. Radiology and Oncology 46 (2012): 207-212.
- Vi?in T, Christensen IJ, Wilhelmsen M, et al. Prognostic and predictive value of cathepsin X in serum from colorectal cancer patients. BMC Cancer 14 (2014): 259.
- Wang J, Chen L, Li Y, et al. Overexpression of cathepsin Z contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. PloS One 6 (2011): 24967.
- Jechorek D, Votapek J, Meyer F, et al. Characterization of cathepsin X in colorectal cancer development and progression. Pathology Research and Practice 210 (2014): 822-829.
- Clark MJ, Homer N, O’Connor BD, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genetics 6 (2010): 1000832.
- Ricard D, Idbaih A, Ducray F, et al. Primary brain tumours in adults. Lancet (London, England) 379 (2012): 1984-1996.
- Biedler JL, Helson L, Spengler BA. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Research 33 (1973): 2643-2652.
- Dawson PJ, Wolman SR, Tait L, et al. MCF10AT: a model for the evolution of cancer from proliferative breast disease. The American Journal of Pathology 148 (1996): 313.
- Soule HD, Vazquez J, Long A, et al. A human cell line from a pleural effusion derived from a breast carcinoma. Journal of the National Cancer Institute 51 (1973): 1409-1416.
- Song Y, Luo Q, Long H, et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Molecular Cancer 13 (2014): 65.
- Amoêdo ND, Valencia JP, Rodrigues MF, et al. How does the metabolism of tumour cells differ from that of normal cells. Bioscience Reports 33 (2013): 865-873.
- Porporato PE, Dhup S, Dadhich RK, et al. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Frontiers in Pharmacology 2 (2011): 49.
- Díaz-Ramos A, Roig-Borrellas A, García-Melero A, et al. α-Enolase, a multifunctional protein: its role on pathophysiological situations. Journal of Biomedicine & Biotechnology 2012 (2012): 156795.
- Splinter TAW, Verkoelen CF, Vlastuin M, et al. Distinction of two different classes of small-cell lung cancer cell lines by enzymatically inactive neuron-specific enolase. British Journal of Cancer 66 (1992): 65-69.
- Loja T, Chlapek P, Kuglik P, et al. Characterization of a GM7 glioblastoma cell line showing CD133 positivity and both cytoplasmic and nuclear localization of nestin. Oncology Reports 21 (2009): 119-127.
- Pišlar AH, Kos J. C-terminal peptide of γ-enolase impairs amyloid-β-induced apoptosis through p75 (NTR) signaling. Neuromolecular Medicine 15 (2013b): 623-635.
- Hao X, Sun B, Hu L, et al. Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer 100 (2004): 1110-1122.
- Miremadi A, Pinder SE, Lee AHS, et al. Neuroendocrine differentiation and prognosis in breast adenocarcinoma. Histopathology 40 (2002): 215-222.
- Nägler DK, Lechner AM, Oettl A, et al. An enzyme-linked immunosorbent assay for human cathepsin X, a potential new inflammatory marker. Journal of Immunological Methods 308 (2006): 241-250.
- Luo H, Shen K, Sun H, et al. Correlation study between serum neuro- specific enolase and gastric and colorectal cancers. Medicine 99 (2020): 19796.
- Pan X, Wu H, Chen G, et al. Prognostic Value of Enolase Gene Family in Colon Cancer. Medical Science Monitor. International Medical Journal of Experimental and Clinical Research 26 (2020): 922980.
- Puzer L, Cotrin SS, Cezari MHS, et al. Recombinant human cathepsin X is a carboxymonopeptidase only: a comparison with cathepsins B and L. Biological Chemistry 386 (2005): 1191-1195.
- Altman DG, McShane LM, Sauerbrei W, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Medicine 9 (2012): 1001216.
Supplementary Information
Supplementary Information 1
Calibration curve of the uncleaved gamma-enolase ELISA.
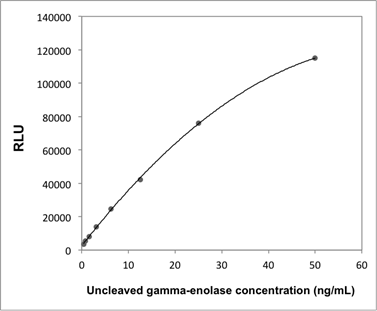
Abbreviation: RLU; relative light unit.
Supplementary Information 2
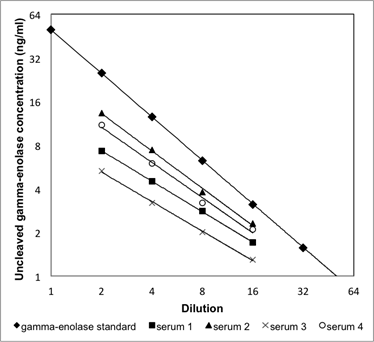
Linearity of the uncleaved gamma-enolase ELISA. Four serum samples were serially diluted (from 1:2 to 1:16) and linearity was checked by comparing to the linearity of the calibration curve. Dilutions of four serum samples showed a linear response, parallel to the calibration curve.
Supplementary Information 3
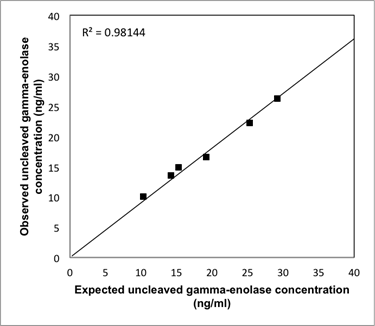
Analytical recovery of the uncleaved gamma-enolase ELISA. Three different amounts of human native gamma-enolase were added to two serum samples from patients with CRC with known amounts of gamma-enolase. The results are shown as observed (measured) vs. expected uncleaved gamma-enolase concentration.
