A Functional Pre-Screening Platform for Identifying Points of Vulnerability in the Cell Death Map of Human Melanoma Tumors
Article Information
Naama Pnina Dekel-Bird1, Shani Bialik1, Orit Itzhaki2, Tomer Meir Salame3, Naama Yaeli-Slonim1, Vered Levin-Salomon1, Santosh Kumar Dasari1,4, Michal Besser2,5,6, Adi Kimchi1*
1Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel
2Ella Lemelbaum Institute for Immuno Oncology, Sheba Medical Center, Ramat Gan, Israel
3Life Sciences Core Facilities, Weizmann Institute of Science, Rehovot, Israel
4Current affiliation: Department of Gynecologic Oncology and Reproductive Medicine, MD Anderson Cancer Center, Houston, TX, USA
5Department of Clinical Microbiology and Immunology, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel
6Wohl Institute for Translational Medicine, Sheba Medical Center, Ramat Gan, Israel
*Corresponding Author: Adi Kimchi, Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel.
Received: 15 November 2021; Accepted: 07 December 2021; Published: 27 January 2022
Citation:
Dekel-Bird NP, Bialik S, Itzhaki O, Salame TM, Yaeli-Slonim N, Levin-Salomon V, Dasari SK, Besser M, Kimchi A. A Functional Pre-Screening Platform for Identifying Points of Vulnerability in the Cell Death Map of Human Melanoma Tumors. Journal of Cancer Science and Clinical Therapeutics 6 (2022): 39-60.
Share at FacebookAbstract
Targeted drug therapy in melanoma patients carrying the BRAF V600E mutation provides temporary remission, often followed by relapse due to acquired drug resistance. Here we propose a functional approach to circumvent drug resistance by applying a personalized prescreening platform that maps points of vulnerability in each tumor, prior to drug treatment. This platform applies siRNAs targeting 81 apoptosis, autophagy and programmed necrosis genes in patient tumor cell cultures, identifying genes whose targeting maximizes cell killing by short-term BRAF inhibition. Melanoma tumors displayed large heterogeneity in the number and identities of soft-spots, providing different tumor-specific functional death signatures. The soft-spots were targeted by replacing functional siRNAs with small compound inhibitors for long-term treatment in combination with vemurafenib. This strategy reduced the number of drug-tolerant persister cells surviving treatment, and most importantly, the number of drug-resistant foci. Thus, prescreening melanoma tumors for soft-spots within the cell death network may enhance targeted drug therapy before resistance emerges, thereby reducing the odds of developing drug-resistant mutations, and preventing tumor relapse.
Keywords
BRAF; Combinatorial drug therapy; Melanoma; Precision medicine; Programmed cell death
BRAF articles; Combinatorial drug therapy articles; Melanoma articles; Precision medicine articles; Programmed cell death articles
BRAF articles BRAF Research articles BRAF review articles BRAF PubMed articles BRAF PubMed Central articles BRAF 2023 articles BRAF 2024 articles BRAF Scopus articles BRAF impact factor journals BRAF Scopus journals BRAF PubMed journals BRAF medical journals BRAF free journals BRAF best journals BRAF top journals BRAF free medical journals BRAF famous journals BRAF Google Scholar indexed journals Combinatorial drug therapy articles Combinatorial drug therapy Research articles Combinatorial drug therapy review articles Combinatorial drug therapy PubMed articles Combinatorial drug therapy PubMed Central articles Combinatorial drug therapy 2023 articles Combinatorial drug therapy 2024 articles Combinatorial drug therapy Scopus articles Combinatorial drug therapy impact factor journals Combinatorial drug therapy Scopus journals Combinatorial drug therapy PubMed journals Combinatorial drug therapy medical journals Combinatorial drug therapy free journals Combinatorial drug therapy best journals Combinatorial drug therapy top journals Combinatorial drug therapy free medical journals Combinatorial drug therapy famous journals Combinatorial drug therapy Google Scholar indexed journals Melanoma articles Melanoma Research articles Melanoma review articles Melanoma PubMed articles Melanoma PubMed Central articles Melanoma 2023 articles Melanoma 2024 articles Melanoma Scopus articles Melanoma impact factor journals Melanoma Scopus journals Melanoma PubMed journals Melanoma medical journals Melanoma free journals Melanoma best journals Melanoma top journals Melanoma free medical journals Melanoma famous journals Melanoma Google Scholar indexed journals Precision medicine articles Precision medicine Research articles Precision medicine review articles Precision medicine PubMed articles Precision medicine PubMed Central articles Precision medicine 2023 articles Precision medicine 2024 articles Precision medicine Scopus articles Precision medicine impact factor journals Precision medicine Scopus journals Precision medicine PubMed journals Precision medicine medical journals Precision medicine free journals Precision medicine best journals Precision medicine top journals Precision medicine free medical journals Precision medicine famous journals Precision medicine Google Scholar indexed journals Programmed cell death articles Programmed cell death Research articles Programmed cell death review articles Programmed cell death PubMed articles Programmed cell death PubMed Central articles Programmed cell death 2023 articles Programmed cell death 2024 articles Programmed cell death Scopus articles Programmed cell death impact factor journals Programmed cell death Scopus journals Programmed cell death PubMed journals Programmed cell death medical journals Programmed cell death free journals Programmed cell death best journals Programmed cell death top journals Programmed cell death free medical journals Programmed cell death famous journals Programmed cell death Google Scholar indexed journals Immunotherapy articles Immunotherapy Research articles Immunotherapy review articles Immunotherapy PubMed articles Immunotherapy PubMed Central articles Immunotherapy 2023 articles Immunotherapy 2024 articles Immunotherapy Scopus articles Immunotherapy impact factor journals Immunotherapy Scopus journals Immunotherapy PubMed journals Immunotherapy medical journals Immunotherapy free journals Immunotherapy best journals Immunotherapy top journals Immunotherapy free medical journals Immunotherapy famous journals Immunotherapy Google Scholar indexed journals targeted therapy articles targeted therapy Research articles targeted therapy review articles targeted therapy PubMed articles targeted therapy PubMed Central articles targeted therapy 2023 articles targeted therapy 2024 articles targeted therapy Scopus articles targeted therapy impact factor journals targeted therapy Scopus journals targeted therapy PubMed journals targeted therapy medical journals targeted therapy free journals targeted therapy best journals targeted therapy top journals targeted therapy free medical journals targeted therapy famous journals targeted therapy Google Scholar indexed journals drug-tolerant persister cells articles drug-tolerant persister cells Research articles drug-tolerant persister cells review articles drug-tolerant persister cells PubMed articles drug-tolerant persister cells PubMed Central articles drug-tolerant persister cells 2023 articles drug-tolerant persister cells 2024 articles drug-tolerant persister cells Scopus articles drug-tolerant persister cells impact factor journals drug-tolerant persister cells Scopus journals drug-tolerant persister cells PubMed journals drug-tolerant persister cells medical journals drug-tolerant persister cells free journals drug-tolerant persister cells best journals drug-tolerant persister cells top journals drug-tolerant persister cells free medical journals drug-tolerant persister cells famous journals drug-tolerant persister cells Google Scholar indexed journals mutations articles mutations Research articles mutations review articles mutations PubMed articles mutations PubMed Central articles mutations 2023 articles mutations 2024 articles mutations Scopus articles mutations impact factor journals mutations Scopus journals mutations PubMed journals mutations medical journals mutations free journals mutations best journals mutations top journals mutations free medical journals mutations famous journals mutations Google Scholar indexed journals tumor relapse articles tumor relapse Research articles tumor relapse review articles tumor relapse PubMed articles tumor relapse PubMed Central articles tumor relapse 2023 articles tumor relapse 2024 articles tumor relapse Scopus articles tumor relapse impact factor journals tumor relapse Scopus journals tumor relapse PubMed journals tumor relapse medical journals tumor relapse free journals tumor relapse best journals tumor relapse top journals tumor relapse free medical journals tumor relapse famous journals tumor relapse Google Scholar indexed journals
Article Details
1. Introduction
Immunotherapy and targeted therapy are the two main therapeutic arms currently available for treating melanoma. While over recent years, immune checkpoint inhibitors have been a breakthrough in the treatment of patients with advanced melanoma, a substantial portion of patients are refractory to or progress after treatment [1,2]. A major difficulty also persists for targeted therapy, which is based on drugs targeting the Mitogen-Activated Protein Kinase (MAPK) signaling pathway that dominates melanoma tumors. In 50% and 20% of melanoma patients, the MAPK pathway is stimulated by mutations in BRAF or NRAS, respectively [3]. Specific drugs targeting the most abundant BRAF V600E mutation (e.g., vemurafenib [4]), MEK (MAP2K) inhibitors [5] and ERK1/2 (MAPK1/3) inhibitors [6] have been developed and used in the clinic. While very effective during the first months of treatment, in a significant fraction of patients, positive tumor responses are followed by a relapse due to the development of acquired drug resistance. Combining MEK inhibitors with BRAF inhibitors from the start of treatment delays the time until relapse, but as in the case of mono-therapy, does not prevent it [7]. While overall, current treatment strategies have improved 5-year survival rates of patients with metastatic melanoma from 5-10% to around 50%, there is still great need for improving the efficacy of standard target therapy to raise survival rates.
Acquired drug resistance develops from persister cells, sub-populations of cancer cells that maintain viability under continuous drug treatment [8-12]. These cells do not harbor inheritable genetic or stable epigenetic modifications, and their drug tolerance phenotype is reversible. They may replicate at low frequency or remain quiescent in the continuous presence of drug. The persister cells are a dangerous cellular reservoir from which acquired mutations that bypass the effect of the drug and confer drug resistance develop during prolonged treatments. Much effort has been expended in recent years in identifying these mutations, and in designing accordingly a second front of drug treatment that targets the new mutations. For example, acquired resistance to BRAF inhibitors has been attributed to activating mutations in MEK1 [13] or MEK2 [14], mutated BRAF splice variants with increased kinase activity [15] and overexpression of mutated BRAF itself. Unfortunately, at this stage, tumors are increasingly refractory even to targeted treatment. Yet due to the multiplicity of such mutations, and their consequent unpredictable nature, mechanisms of potential resistance cannot be targeted at earlier stages.
Here we suggest an alternative way to address the problem of acquired drug resistance by identifying points of vulnerability in melanoma tumors at the start of vemurafenib treatment, the targeting of which would reduce the number of persisters and as a consequence, the odds of developing drug resistant mutations. It is based on the premise that the suppression of the MEK/ERK pathway by BRAF inhibitors can be wired in different ways to various molecular mechanisms that ultimately reduce tumor cell number, i.e. different pathways that control the life and death decisions of a cell, such as apoptosis, necrosis, and autophagy [16-20]. We use a Programmed Cell Death (PCD) siRNA library to identify points-of-vulnerability within these various PCD pathways in melanoma tumors at the start of vemurafenib treatment, and assign a tumor-specific cell death signature. Subsequent targeting of the soft-spots identified in this manner significantly reduced the number of persister cells, and most importantly, attenuated the subsequent long-term development of drug resistant foci.
2. Materials and Methods
2.1. Reagents, cell culture and transfections
All chemical reagents were purchased from Sigma-Aldrich unless otherwise stated. Primary melanoma lines were generated from tumor biopsies of metastatic melanoma patients, as detailed in Supplementary Table S1, enrolled in a phase II TIL ACT trial at the Sheba Medical Center. Patients signed an informed consent approved by the Israeli Ministry of Health (Helsinki approval no. 3518/2004, NCT00287131), which allowed enrollment in the clinical trial and the use of excess cell material for research purpose. Fragmentation, enzymatic digestion and tissue remnant culture techniques were used to isolate melanoma cells from surgically resected metastatic lesions to generate primary melanoma cell lines [22]. These cell lines, which tested negative for mycoplasma at thawing, were maintained in T2 medium (RPMI, 10% FBS (HyClone, Thermo Scientific, SH3010903), 1mM sodium pyruvate, 25mM HEPES, Pen/Strep, L-Glutamine (all from Biological Industries). The cells were treated with vemurafenib at the indicated concentrations (Selleckchem, S1267), 0.5µM or 1µM S63845 (ApexBio, A8737-5), 0.5µM or 1µM ABT-737 (Santa Cruz, sc-207242), 20µM HCQ (Sigma, C6628, H0915), 5-10nM bafilomycin A1 (LC-Laboratories, B-1080), 0.5µM or 1µM venetoclax (Medchemexpress, HY-15531) and 0.5µM or 1µM A-1331852 (Medchemexpress, HY-19741). For transient transfections of siRNA, 25nM siGNOME siRNA pool (Dharmacon), or a total of 50nM for double KD, was mixed with transfection reagent Dharmafect1 (Dharmacon) and added to the cells 24h post plating.
2.2. Cell viability and death assays
Cell viability was measured using luminescence based CellTiter-Glo assay according to the manufacturer’s instructions (Promega, G7572). Luminescence was read in a microplate luminometer (TECAN, Tecan Trading AG). For calcein/PI assays, 2.5x105 cells were grown in 6-well plates and 24h post treatment, 1µM Calcein AM (Life Technologies #C3099) and 1.5µM PI (Sigma, P4864) were added. The plates were then incubated for 30 min at 37°C and live and dead cells were viewed by fluorescence microscopy (Olympus BX41 or Olympus IX73 microscopes) equipped with 10x or 20x objectives, and digital images obtained with DP72 CCD or DP73 cameras (Olympus) using CellSens Standard software (Olympus). For establishing the platform, we used a custom designed PCD siRNA library (Dharmacon) comprising 81 siRNAs targeting apoptotic, necroptotic and autophagic genes. These were arranged randomly in two 96 well white flat bottom plates (Greiner, 655083) with several control siRNAs scattered among them (25 nmol each). Eight siRNAs were placed on both plates as internal standards to enable plate comparison. The melanoma cell lines were then reverse transfected in the 96 well plates (5000 cells/well). 24h post-transfection the medium was replaced with fresh medium, and 48h post transfection vemurafenib (10µM) was added to the treated cells and DMSO to the untreated cells. After 24h of treatment, cell viability was measured by the CellTiter-Glo assay. Each cell line was assessed in triplicate, on different days.
Analysis of the functional death signature was done using R as follows: The value of the 8 genes that were represented on both plates was the average of the two technical repetitions. From the raw values, the value of blank cells was subtracted, then the average value of the si-controls from each plate was used to normalize the value of each siRNA. The si-control value was set as 1. The normalized values from each repetition were averaged. The ratio of the ln average of the treated divided by the ln average of the untreated value was the mean ratio that was used to determine the change in the viability. A value of 0 refers to no change in the viability when knocking down the gene between the treated and the untreated samples, positive and negative values refer to increased and decreased viability, respectively. Hits were those knock-downs that passed two criteria: the change in the viability had to be significant (p<0.05) in comparison to the si-control according to a Dunnet test, and in comparison to the untreated KD with the same siRNA by student T-test. Both p-values appear in Supplementary Table S2.
2.3. Cell cycle analysis
For cell cycle analysis, 2x106 cells were pulsed with 10µM BrdU (Sigma, B5002) for 2-4h and both detached cells and trypsinized attached cells were collected. Cells were fixed in cold ethanol overnight at 4°C, incubated for 30 min in Denaturation Solution (2N HCl/Triton X-100 in PBS 1X), washed in Neutralization Solution (0.1 M Na2B4O7, pH 8.5) and incubated 1h with Antibody Solution (PBS 1X, 0.5% Tween 20, 1% BSA, 1µg/100µl anti-BrdU FITC antibody (eBioscience, cat#11-5071-41). Cells were then resuspended in PBS containing 50 µg/ml PI and 50 µg/ml RNase A. Flow cytometry analysis was performed on a BD LSRII instrument (BD Immunocytometry Systems) equipped with 355-, 407-, 488-, 561- and 633-nm lasers, controlled by BD FACS Diva software v8.0.1 (BD Biosciences), at the Weizmann Institute of Science's Flow Cytometry Core Facility. FITC was detected by excitation at 488 nm and collection of emission using 505 longpass (LP) and 525/50 Bandpass (BP) filters. PI was detected by excitation at 561 nm and collection of emission using 600 LP and 610/20 BP filters. Further analysis was performed using FlowJo software v10.2 (Tree Star).
2.4. Protein analysis
Cells were lysed in PLB buffer (10mM NaPO4 pH 7.5, 5mM EDTA, 100mM NaCl, 1% Triton X-100, 1% Na deoxycholate, 0.1% SDS) supplemented with 10μl/ml 0.1M PMSF and 1% protease and phosphatase inhibitor cocktails. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes, which were incubated with antibodies against ERK (cat#M5670), pERK (cat#M8159), vinculin (cat#V9131) from Sigma; FIP200 (cat#12436), Atg12 (cat#4180P), BIM (cat#2819), cleaved caspase-3 (cat#C-9664) from Cell Signaling; or Mcl-1 (Santa Cruz, sc-20679). Detection was done with either HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies (Jackson ImmunoResearch, cat#211-032-171, 111-035-144), followed by enhanced chemiluminescence (EZ-ECL, Biological Industries Israel Beit-Haemek Ltd.)
2.5. Statistics
The statistical significance of differences between means was assessed either by standard Student’s T-test or by one-way ANOVA followed by Tukey’s post hoc test, as stated in the Figure legends. Values with p<0.05 were considered significant. Statistical analysis was done using GraphPad Prism version 7.04 (La Jolla California, USA) or for the large screen viability output, R 3.3.1 (2016-06-21), with the nparcomp, multcomp and the pheatmap packages.
3. Results
3.1. Patient-derived melanoma cell lines carrying the BRAF V600E mutation show heterogeneous responses to vemurafenib treatment
A cohort of 12 early passage melanoma cell cultures were established from individual patient tumors carrying the V600E mutation of BRAF. Tumour cell lines were derived from metastatic patients before treatment with the clinically approved BRAF inhibitors (Supplementary Table S1). Despite the common activating BRAF mutation, there was a wide heterogeneity in the response of the cell lines to 24h treatment with 10µM vemurafenib; the extent of reduction in cell number/viability, as assessed by the CellTiter-Glo assay, ranged from 20-80% in 11/12 cell lines, and one cell line showed no change (Figure 1A). Western blotting indicated a decline in phosphorylation of ERK, the downstream target of BRAF, in all cell lines except for line 13, indicating that the drug was effective in inhibiting BRAF in the other 11 cell lines (Figure 1B). Notably, in line 13, the high basal ERK phosphorylation levels were even further increased by the drug, suggesting that ERK phosphorylation is upregulated in this tumor by some second unknown mechanism independent of the V600 BRAF mutation. BIM (BCL2L11) protein levels were elevated in all drug-sensitive cell lines, except for line 43, a consequence of its stabilization upon reduced phosphorylation by ERK [23].
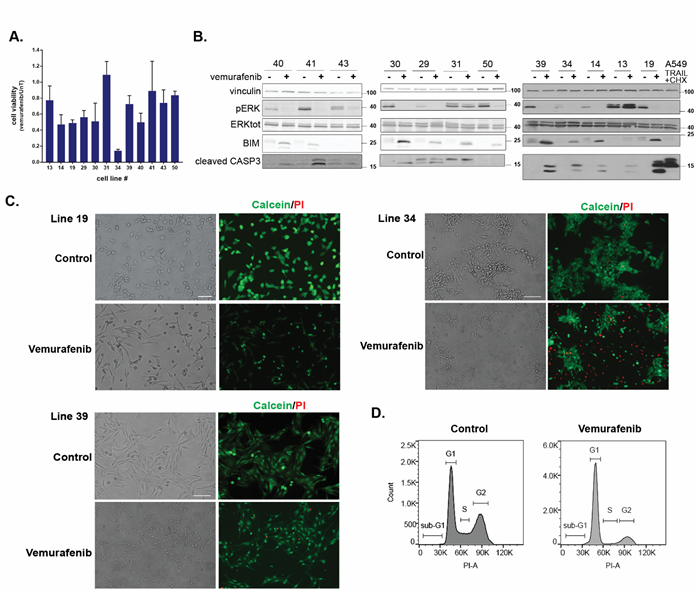
Figure 1: Patient-derived melanoma cell lines show heterogeneous responses to vemurafenib treatment. A. Quantification of cell viability by CellTiter-Glo assay in response to 10μM vemurafenib treatment for 24h. CellTiter-Glo reads of the untreated cell lines (UnT) were normalized to a value of 1 and the y-axis values indicate the relative decrease in response to vemurafenib treatment. Shown are the mean±SD of triplicates, for each cell line. B. Western blots monitoring the responses of the 12 melanoma cell lines to vemurafenib treatment showing: ERK phosphorylation, Bim expression levels and cleaved Caspase-3 levels. A549 cells treated with TRAIL and cycloheximide were used as a positive control for strong caspase-3 activation during apoptosis. Lysates were run on three different blots as the cell lines became available for analysis, and blots were cut into appropriate strips for simultaneous probing with antibodies to proteins of different sizes. Antibodies to vinculin were used as loading control on all blots. For ERK detection, blots were first probed for phospho-ERK, stripped and reprobed with antibodies to total ERK. For the blots of cell lines shown at left and right, the same lysate was run on two separate gels to examine proteins with overlapping sizes, for the middle set of cell lines, only 1 gel was run, and the slice of membrane used to detect Caspase-3 was stripped and reprobed with anti-Bim antibodies. C. Representative phenotypic response to vemurafenib of cell lines 19, 34 and 39. Cells were treated with vemurafenib for 24h, and then stained with calcein AM (green, stains live cells) and propidium iodide (PI) (red, stains cells with permeabilized membranes) and viewed at 10x (lines 34, 39) or 20x magnification (line 19). Greyscale versions of single filter channels for calcein and PI can be seen in Supp. Fig. S1A. D. Cell cycle distribution of line 39 treated with 5μM vemurafenib for 24h and stained with PI. 50,000 cells were recorded for each sample. Shown is a representative experiment of three independent repetitions. The X-axis corresponds to PI staining and the Y-axis to cell number.
Phenotypic analysis following staining with calcein-AM/PI indicated that the reductions in CellTiter-Glo values were caused by at least three different cell responses during the first 24h of drug treatment (Figure 1C and Supplementary Figure S1A). A high incidence of dying round, shrunken cells was observed in some cell lines, such as lines 34 and 19. This was associated with membrane permeabilization, as documented by the strong PI staining, in line 34, but not in line 19, suggesting that line 34 cells die by programmed necrosis, whereas line 19 cells undergo an early apoptotic response. In contrast, in cell line 39, the abundance of cells with a death related phenotype at the 24h time-point was low, suggesting that the reductions in the CellTiter-Glo values mostly reflect proliferation arrest rather than cell death. Flow cytometry cell cycle analysis of line 39 confirmed the microscopic observations. At 24h post drug treatment, the cells accumulated in G1, at the expense of S and G2 phases (Figure 1D), and had reduced BrdU incorporation (Supplementary Figure S1B), indicative of a block in cell cycle progression. This initial morphological survey documents heterogeneity in drug responses to vemurafenib among patient-derived cell lines ranging from cytostatic to different types of cytotoxic responses.
Molecular analysis of the death response by western blotting was consistent with the observed phenotypic heterogeneity (Figure 1B). Caspase-3 (CASP3) was cleaved to its activated products to varying extents in approximately half of the cell lines, with line 19 showing the most robust cleavage, similar to the positive control of A549 cells treated with TRAIL and cycloheximide (Figure 1B). The remaining cell lines either failed to activate caspase-3, or did not show enhanced caspase-3 cleavage in response to the drug compared to control. This was despite the suppression of the MAPK pathway and stabilization of the pro-apoptotic BH3-only protein BIM in all but one of these cell lines. This suggests that BIM stabilization is not sufficient in itself to promote caspase-activating pathways. In these latter cell lines, caspase-independent pathways are limiting the growth and/or cell viability, at least during the first 24h of drug treatment. Altogether these initial experiments underscore the large heterogeneity that exists in the phenotypic and molecular responses to vemurafenib treatment in melanoma.
3.2. Mapping points-of-vulnerability in the programmed cell death map of patient-derived melanoma cell lines
The less than optimal and variable death responses to vemurafenib within the melanoma cohort suggest that strategies to further reduce cell viability should be “personalized” to activate the most effective cell death pathway in each cell line. To this end, based on our previously curated PCD map (Supplementary Figure S2A), we generated a siRNA-based pre-screening platform for studying the fidelity of various cell death pathways in individual melanomas, to identify soft-spots in the cell death machinery that can be targeted for maximal killing in each tumor. The detailed outline of the platform is depicted in Figure 2A. Briefly, a custom designed PCD siRNA library (Dharmacon) comprising 81 siRNAs targeting apoptosis, autophagy and programmed necrosis genes were arranged randomly in two 96-well plates with several control siRNAs scattered among them (Supplementary Figure S2B). The siRNAs were reverse transfected into the melanoma cell lines, and cell viability in response to vemurafenib was then assessed for individual knock-down cells using the CellTiter-Glo assay. For each cell line, the screen was done in triplicate (Supplementary Table S2). These results were converted into heat maps depicting the varying levels of viability for each KD cell line, and were organized in a combined global heat map display for the 12 cell lines (Figure 2B). A functional hit was defined as a statistically significant siRNA-mediated effect that moderated the vemurafenib-induced reduction in cell viability, by either enhancing cell death (positive hits, red bars of varying intensity in Figure 2B) or by decreasing cell death (negative hits, blue bars of varying intensity in Figure 2B). By determining the contribution of each gene to the death response to vemurafenib in this manner, the overall ‘functional death signature’ of each tumor was characterized, highlighting the specific death pathways that are functionally active. The positive hits identified within these pathways are the ‘soft-spots’ that can be targeted for enhanced cell killing.
The global overview of the combined heat maps of the 12 cell lines showed a large heterogeneity in the number and position of the soft-spots within the three modules of the PCD map (Figure 2B and Supplementary Table S3). The average number of soft-spots was 6. However some cell lines (19, 30, 34, 39, 40, 43) were super-sensitive, with up to 15 soft-spots scattered among the PCD modules, mostly targeting different genes along the PCD pathways in each cell line, with little overlap (see Supplementary Figure S3 for positioning of the hits in the PCD map in lines 19, 34 and 39). The other cell lines displayed fewer soft-spots. More resistant cell lines (i.e. 13, 29 and 31) each displayed a single soft-spot at different positions in the PCD map, and cell line 41 had no detectable soft-spots.
The knock-down results were confirmed by independent validation assays in 2 cell lines with multiple hits. In cell line 34, knock-down of ATG5 reduced its mRNA expression and the steady state levels of the ATG5-ATG12 conjugate (Supplementary Figure S4 A,B), confirming that the ATG5 siRNA is effective in limiting the conjugation process with ATG12, a step that is critical for autophagosome maturation. FIP200 protein levels and DAPK3 mRNA levels were also reduced by the corresponding siRNAs (Supplementary Figure S4A,B). Notably, the knock-down of ATG5 or FIP200 did not affect the upstream dephosphorylation of ERK and consequent stabilization of BIM in response to vemurafenib treatment, suggesting a downstream effect (Supplementary Figure S4B). Also their knock-down did not accelerate the low levels of caspase-3 activation by vemurafenib characteristic of this cell line, implying that the increased cell lethality that resulted from autophagy perturbation did not involve the indirect activation of the apoptotic module. As shown in Figure 3A, the KD of ATG5, DPAK3 and FIP200, and also ATG12 and C-FLIP (CFLAR) (the other soft-spots identified in this cell line) reduced the overall cell viability in response to vemurafenib, similar to the original screen, thus validating the screen results for this cell line.
Cell line 39 had 6 hits (Figure 2B and Supplementary Table S3, Supplementary Figure S3C); validation assays were performed for MCL1 and ATG5 siRNAs. The cell death responses of the KD in combination with vemurafenib were similar to those observed in the large-scale siRNA screen (Figure 3B). As in cell line 34, the KDs did not interfere with upstream dephosphorylation of ERK and stabilization of BIM as a response to vemurafenib treatment (Supplementary Figure S4C). Notably, the KD of anti-apoptotic MCL1 increased cleaved caspase-3. Similarly, and in contrast to line 34, ATG5 KD also led to enhanced caspase activation. The effects of both KDs were partially rescued by the caspase inhibitor Q-VD (Figure 3C). This suggests that the knock-down of ATG5 in combination with vemurafenib augments activation of the apoptosis pathway, supporting functional duality. Interestingly, the combined knock-down of ATG5 and MCL1 had an additive effect (Figure 3C), suggesting that they most likely function via different mechanisms to inhibit apoptosis, and that apoptosis can be enhanced by more than one pathway. The additive effects of double perturbations underscore the importance of unveiling several potential non-redundant soft-spots in each tumor.
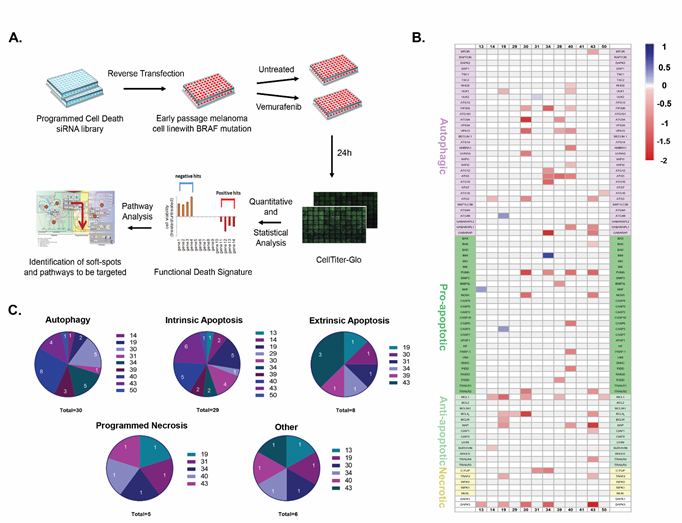
Figure 2: Mapping points of vulnerability in the programmed cell death map in each tumor. A. Schematic of the siRNA-based screening platform used for delineating the functional death signature of each cell line. Each experiment was done in triplicate. B. Combined heatmap of the hits for all cell lines. Red corresponds to positive hits (soft-spots), blue to negative hits, and genes whose knock-down did not statistically affect the responses to the drug are in background color (white or light grey). Genes are grouped according to cell death sub-modules; DAPK1 and DAPK3, multifunctional regulators of various pathways that are not part of the basic cell death machinery, are not included in any specific sub-module. C. Pie charts indicating distribution and number of soft-spots per cell line, within each cell death sub-module (autophagy, intrinsic and extrinsic apoptosis and programmed necrosis; “other” refers to the DAPK family, specifically DAPK3, the only soft-spot within this group).
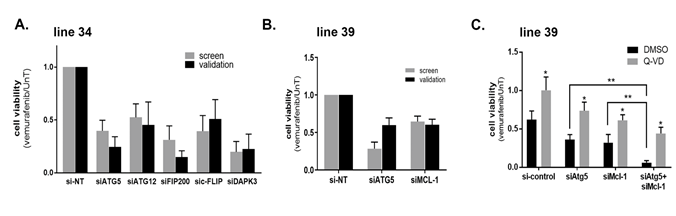
Figure 3: Validating points of vulnerability in the programmed cell death map in each tumor. A.-C. Cell viability in response to 10μM vemurafenib was tested 48h after transfection with the indicated siRNAs or siRNA combinations using the CellTiter-Glo assay in line 34 (A) or line 39 (B,C). In C, 50mM Q-VD, or DMSO as control, were added. The values were first normalized to si-control knock-down cells, and then the ratio of drug treated to untreated calculated, with the ratio of the si-control knock-down cells set as 1. Data represents mean±SD of three replicate experiments. In C, statistical significance of the difference across siRNA manipulations was assessed using one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test, ** p <0.01. Statistical significance of Q-VD vs. DMSO treatment was determined by two-tailed paired Student’s T-test, * p<0.05.
3.3. Pathway analysis
Several interesting conclusions emerged from the analysis of the functional death signatures of the various cell lines. The most frequent soft-spot was the Ser/Thr kinase DAPK3 (positive hit in 6/12 cell lines) (Figure 2B,C and Supplementary Table S3). In 4 additional cell lines, KD of DAPK3 alone significantly reduced cell viability and the addition of vemurafenib did not enhance cell killing, and therefore it did not pass our definition of a positive hit (Supplementary Table S2).
Not surprisingly, the apoptosis pathway was important for enhancing the cell death response to vemurafenib (Figure 2B,C and Supplementary Table S3); soft-spots corresponding to apoptosis inhibitors from within the intrinsic and extrinsic apoptosis pathways were observed in 10/12 and 5 cell lines, respectively. Importantly, the identities of the soft-spots differed among the cell lines, and mapped to various members of the BCL2 and/or IAP families. The most common positive hit was the anti-apoptotic BCL2 family member MCL1, which was found in 5/12 cell lines. Counterintuitively, in 4 cell lines, KD of PUMA (BBC3) or NOXA (PMAIP1), members of the BH3-only protein subfamily, led to enhanced killing. This suggests that in specific cellular contexts, these proteins may display a function opposite of the canonical pro-apoptotic roles previously reported for BH3-only proteins.
The Bcl-2 family death signatures served as a paradigm to investigate the specificity of the screening platform. To this end, the various knock-downs were replaced with specific inhibitors S63845 for MCL1, A1331852 for BCLXL (BCL2L1), venetoclax for BCL2, and ABT-737, which targets BCL2, BCLXL and BCLW (BCL2L2), in cell lines 14, 19, 39, 40, and 43 (Figure 4A). For each cell line, there was a significant reduction in the viability of the cells after 48h co-treatment with vemurafenib, only upon inhibition of the BCL2 family members that were positive hits in the screen. For example, in line 39, the MCL1 specific inhibitor S63845, but not the BCL2 or BCLXL specific inhibitors, led to an enhanced cell death response to vemurafenib (Figure 4B). S63845 also enhanced cell killing in lines 14 and 19, in which MCL1 was a soft-spot, as in line 39 (Figure 4C, D). In contrast, the MCL1 inhibitor had no additional effect on cell killing in line 43 (Figure 4E), in which MCL1 was not a positive hit, although the BCLXL inhibitor did enhance cell killing. The broad range ABT-737 that does not inhibit MCL1 had no effect on line 39 or line 14, but did reduce cell viability in line 19, in which BCLXL and BCLW were identified as positive hits (Figure 4B-D). Notably, none of the BCL2 family inhibitors had any effect on vemurafenib-induced cell killing in line 40, which showed no soft-spot within the family, and served as the negative control in these experiments (Figure 4F). There was no change in viability from treatment with inhibitors alone (see for example, Supplementary Figure S4D).
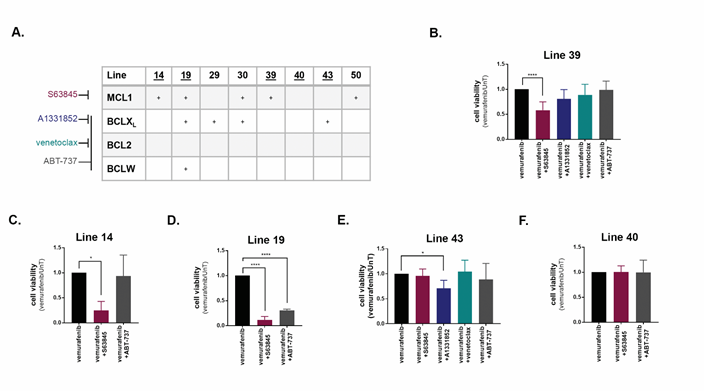
Figure 4: Inhibition of BCL2 family members promotes cell death induced by vemurafenib in a cell-line specific manner. A. Chart indicating positive hits within the anti-apoptotic BCL2 family in the indicated cell lines. Specificity of inhibitors are indicated at left. Underlined cell lines are those in which inhibitors were tested. B.-F. Cell viability of cell lines treated with the combination of 10μM vemurafenib and either 0.5μM (B-D,F) or 1μM (E) of inhibitors of BCL2 family members for 48h. Data represents mean±SD of three replicate experiments. Statistical significance was assessed using ANOVA followed by Dunnett's multiple comparisons test, * p<0.05, **** p<0.0001.
The second module that emerged with multiple soft-spots was autophagy (Figure 2B,C). Clusters of positive hits within the various steps of autophagy were found in 5/12 cell lines, suggesting that autophagy serves as a survival mechanism in these drug-treated melanoma. For example, cell line 34 had 5 autophagy soft-spots (ATG10, ATG12, ATG5, FIP200 (RB1CC1) and GABARAP) (Figure 2B and Supplementary Table S3, Supplementary Figure S3B). Four of these five genes belong to the ubiquitin-like conjugation pathway that mediates LC3B (MAP1LC3B) conjugation to phosphatidylethanolamine (PE), representing the central execution step of autophagy, membrane elongation. In other cells (lines 30, 39, 40, 43), the hits were more widely dispersed among the different steps. ATG5 and FIP200 were the most common hits, found in 3 cell lines each. A second profile of autophagy soft-spots was also observed (Figure 2C): three cell lines (lines 14, 19, 50) had single hits at different positions within the pathway, as opposed to multiple hits throughout the pathway (see for example line 19, Supplementary Figure S3A). In these cell lines, the emergence of lone autophagy proteins as soft-spots suggests that these genes may serve a second cell death function independent of autophagy, a phenomenon that has been documented for several autophagy proteins [21] and is depicted by cross-pathway interactions within our PCD map.
The identification of the cluster of autophagy genes soft-spots in cell line 34 suggested that the autophagy process as a whole was critical for counteracting the cell death responses to vemurafenib. In support of this, vemurafenib and general autophagy inhibitors Hydroxychloroquine (HCQ) or Bafilomycin A, which block fusion of the lysosome with the autophagosome, enhanced the reduction in cell viability compared to vemurafenib alone by 50% in line 34 (Figure 5A). Neither autophagy inhibitor had any effect on cell viability alone (Supplementary Figure S4D). In contrast, cell line 39, which had only 3 sporadic autophagic hits, at least one of which (ATG5) was linked to the apoptosis pathway, did not show enhanced cell killing when treated with HCQ or Bafilomycin A in combination with vemurafenib (Figure 5B). Thus the autophagy pathway is a promising target for increasing vemurafenib-induced death responses only if the siRNA-based pre-screening platform indicates multiple potential soft-spots that cluster within the main autophagy pathway stages.
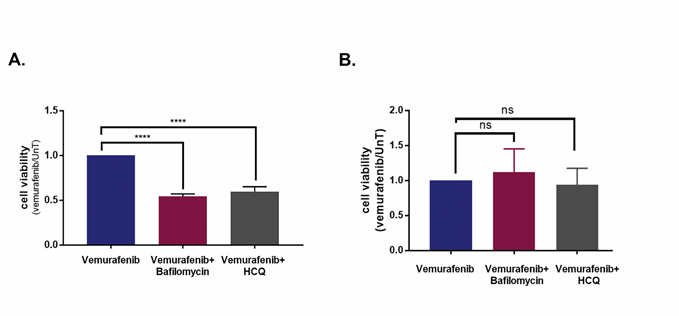
Figure 5: Inhibition of autophagy promotes cell death induced by vemurafenib in a cell-line specific manner. A., B. Cell viability of cell lines 34 (A) and 39 (B), treated with the combination of 10μM vemurafenib and the autophagy inhibitors Bafilomycin A1 (5nM) or HCQ (20μM) for 48h, as assessed by CellTiter-Glo assay. Data represents mean±SD of three replicate experiments. Statistical significance was assessed by ANOVA followed by Dunnett's multiple comparisons test, **** p<0.0001.
3.4. Proof-of-principle: Targeting soft-spots during long term vemurafenib treatment of melanoma cell lines reduces viability of drug tolerant cells and the number of drug resistant foci
The most important clinically relevant question is whether targeting the soft-spots identified in the screen affects long term vemurafenib treatment, and whether it translates into reduced frequency of acquired drug resistance. Since long-term experiments could not be performed with siRNA KDs, KDs were replaced by available drugs for specific siRNAs as a proof-of-principle. Specifically, the additive effects of MCL-1 inhibitor S63845 on the vemurafenib responses in cell line 39 were measured by flow cytometry for DNA content and by CellTiter-Glo for cell viability over a 14d time-course. While treatment with 2mM vemurafenib alone induced G1-arrest during the first 24h (Supplementary Figure S5A), the combination of 2mM vemurafenib and 0.5mM S63845 showed an apoptotic response, as indicated by the elevated sub-G1 population (Figure 6A, Supplementary Figure S5A). At later time-points, the sub-G1 population was always larger in the cells that were treated with the drug combination (Figure 6A), indicative of accelerated apoptosis. The cell viability assay gave similar results; from 1d and up to 14d of treatment, the viability of the cells treated with the drug combination was significantly lower than the viability of cells treated with vemurafenib alone (Figure 6B). Notably, S63845 alone had no detectable effects, as measured both in the cell viability assay (Figure 6B) and by flow cytometry for DNA content (Supplementary Figure S5A).
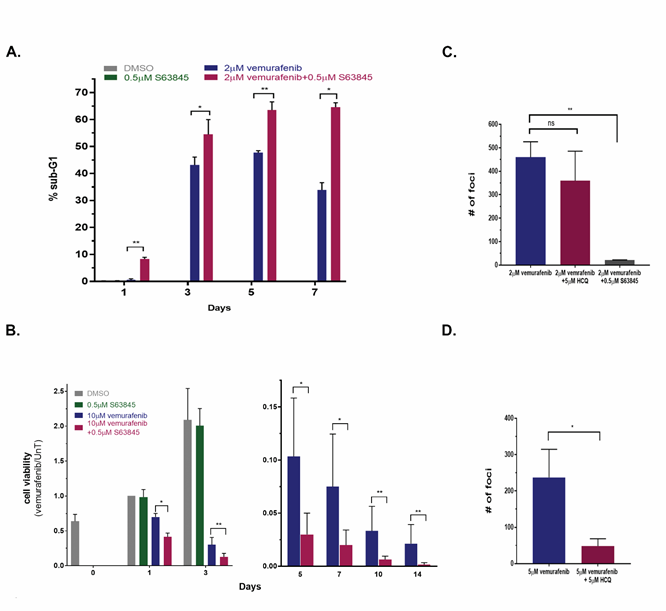
Figure 6: Targeting MCL1 during long term vemurafenib treatment of melanoma cell lines reduces viability of drug tolerant cells and the number of drug resistant foci. A. Line 39 cells were either untreated, or treated with 2μM vemurafenib, 0.5μM S63845 or both for 1, 3, 5 and 7 days. Cells were stained with PI and 50,000 cells per sample were analyzed by FACS in 3 replicates. Shown is the % sub-G1 population. Cells that were treated with DMSO or S63845 alone were only assessed at day 1 since they reached confluence beyond that point (0.18% and 0.28%, respectively). * p<0.05, **p<0.01. B. Cell viability of line 39 left untreated or treated with 10μM vemurafenib, 0.5μM S63845 or both for up to 2 weeks, as assessed by CellTiter-Glo assay. Monitoring of untreated and S63845 treated control cells was discontinued after 3 days, and the data is split accordingly between two graphs with different Y-axis ranges. Values were normalized to day 1 untreated. C, D. 6x106 cells of line 39 (C) or line 34 (D) were plated and treated with 2μM vemurafenib alone or with 5μM HCQ or 0.5μM S63845 for 4 and 5 weeks, respectively. Fresh medium containing the drugs was supplied twice weekly. Individual foci were counted, shown are mean±SD of three independent biological repeats. The treatment with either 0.5μM S63845 or 5μM HCQ alone did not have an effect on the cells, comparable to no treatment, which reached full confluence by 3 days, and thus foci could not be counted within the same time-course and plating conditions. **p<0.01, ns- nonsignificant.
Under continuous vemurafenib treatment, a small fraction of drug tolerant cells was identified displaying the main characteristics of persister cells. That is, the population of surviving cells retrieved and grown after removing the drug at day 10 of treatment retained their sensitivity to vemurafenib similar to the original naïve population (Supplementary Figure S5B). The number of drug tolerant cells that still maintained their viability was further reduced as drug treatment continued (Figure 6B), yet this fraction of cells was not completely eliminated. In contrast, the combination of vemurafenib and MCL1 inhibitor was consistently more effective in reducing cell number during prolonged treatment. After several weeks of continuous exposure to vemurafenib, drug resistant colonies (foci) developed in the plates from the residual cell population. Resistant colonies started to appear in lines 39 and 34 after 35 days of treatment. A cluster of cells was considered to be a focus only when mitotic cells were abundantly identified within it. Most importantly, in line 39, the number of drug resistant foci was reduced 22-fold in the plates simultaneously treated with the MCL1 inhibitor S63845 and vemurafenib, as compared to vemurafenib alone (Figure 6C). In contrast, the combination of vemurafenib and HCQ did not change the number of drug resistant foci, consistent with the inability of the autophagy inhibitor to enhance the initial responses to vemurafenib (Figure 5B) and with the functional death signature of line 39 in the screen (Figure 2B; Supplementary Figure S3C). When testing the combination of vemurafenib with or without HCQ in cell line 34, where the autophagy pathway was identified as a point of vulnerability, the combination treatment reduced foci formation five-fold (Figure 6D). In conclusion, the combination of vemurafenib and drugs that targeted the soft-spots identified by the siRNA screen reduced the number of viable cells under continuous drug treatment, and as a consequence, reduced the number of resistant colonies emerging from this residual cell population.
4. Discussion
Genomic and transcriptomic tools can predict which oncogenes drive a particular cancer, enabling the design of targeted treatment. However they cannot capture the full genetic and epigenetic alterations and complexity of the tumor, and patients that are given targeted therapy solely based on their -omics data tend to relapse [24,25]. Drug combinations are needed to reduce the number of cells that survive below the critical number that enables the development of acquired drug resistance with time. Therefore, functional screening approaches were developed using siRNA and drug libraries in cell lines and organoids obtained from patients to identify novel combinations [26-30]. Montero, et al. [30] developed a functional screen, called Dynamic BH3 Profiling, that can predict if a tumor is primed for apoptosis after certain chemotherapy, based on their response to a panel of BH3 mimetics. In the present study, we undertook a more global approach to profile the broader functional cell death network for personalized precision therapy for melanoma. An siRNA screening platform was used to determine the functional death signature of naïve melanoma tumors carrying V600E BRAF mutation challenged in culture with vemurafenib. This enabled the identification of points-of-vulnerability within the cell death network, referred herein as soft-spots, which can lead to therapeutic strategies using drug combinations tailored to each patient to initiate a strong death response. We have shown that such combinatorial approaches enhance cell death and most importantly, reduce the emergence of resistant clones during long-term growth in culture. Notably, the soft-spots that emerged from the siRNA screen comprised only those targets that were not lethal in the absence of vemurafenib, thus conferring selectivity of the combinatorial treatment towards the tumor cells carrying the BRAF mutation. The ability of this pre-screening platform to map various hits along the apoptosis, autophagy and programmed necrosis pathways provides a broad view of the functional death signature of each tumor, and opens the opportunity for simultaneously targeting multiple hits to further increase the lethal responses, as shown here by the double perturbations of ATG5 and MCL1 in line 39. Clinically, this approach would translate to a personalized treatment protocol aimed at decreasing the tumor size below the threshold that allows the tumor to acquire drug resistance and prevent relapse of the tumor.
Our analysis demonstrates the necessity of mapping the intact cell death pathways that can be activated in each individual tumor. The molecular and phenotypic analysis of the twelve early passage melanoma cell lines derived from individual patient tumors showed different types of responses to vemurafenib treatment, and the siRNA perturbations indicated different functional death signatures in each cell line. This could reflect the fact that the BRAF-ERK1/2 signaling axis can be connected to different cellular processes, and thus its inhibition will have divergent effects on pathways controlling cell growth and death, both caspase-dependent and independent.
It was not surprising that many apoptotic regulators emerged as soft-spots, as vemurafenib treatment has been documented to activate apoptosis in many cases [31,32]. Yet both the diversity of the soft-spots, and the unexpected emergence of some pro-apoptotic proteins among them, underscore the importance of conducting unbiased personalized screens to identify the best means of activating the apoptosis module during vemurafenib treatment. MCL1 and BCLXL were the BCL2 family members present as the most frequent soft-spots, consistent with other studies that have documented high levels of MCL1 and BCLXL in melanoma [33-36] and that have shown that combination treatment of BRAF inhibitor with BH3-mimetics was effective in melanoma cell lines [37,38]. However, choosing the right inhibitor that would be most effective is still a challenge; the analysis presented here shows that different melanomas respond differently to the range of BH3 mimetics currently available. The siRNA pre-screening platform can help to determine when and which BH3-mimetic should be administered.
In addition to apoptosis, autophagy was also found to play a role in the survival of melanoma cell lines. Successful combination of inhibitors from the MAPK pathway with autophagy blockers has been documented in several cancers, such as brain tumors [39], pancreatic cancer [40] and melanoma [16,41,42]. This led to the suggestion that autophagy helps cells cope with the stress induced by drug treatment, and therefore blocking autophagy removes this coping mechanism and enhances the death response. Most of the autophagic hits fell within the ubiquitin-like conjugation pathway that executes the membrane elongation steps of autophagy. Upstream regulators were less prominent; this may be due to the abundance of signaling pathways that converge on the initiation stages of autophagy, redundancy among these regulators (e.g. ULK1 and ULK2), the multiple sources for phagophore membrane initiation, and/or the presence of non-canonical signaling pathways that can occur independently of the VPS34 (PI3KC3) PI3K lipid kinase and ULK1 complexes. The ubiquitin-like conjugation step, in contrast, may serve as a limiting process for the continuation of autophagy. Of note, autophagy inhibitors in use in the clinic to date, such as chloroquine (CQ) and HCQ, are not specific for the autophagy process. Pre-clinical attempts to develop more specific inhibitors have focused on kinases such as VPS34 [43]. Our study, however, indicates that these upstream targets may not be rate-limiting and thus not the best candidates for blocking this survival pathway in melanoma, but rather, drug design should focus on the ubiquitin-like conjugation step. As in the case of the BCL2 family inhibitors, the cell death signature data indicates that inhibiting autophagy is not effective in all melanoma tumors; our study suggests that pre-screening tumor responses to vemurafenib can predict the effectiveness of autophagic inhibitors as a viable co-treatment strategy.
Another prominent hit was DAPK3, detected as a soft-spot in half of the cell lines tested. DAPK3 is a member of the death-associated protein kinase family. It was found to have roles in induction of both apoptosis [44-46] and autophagic cell death [47]. On the other hand, DAPK3 was also found to be important for tumor survival [48-50], which contradicts the well-known tumor suppressive properties of the DAPK family and their ability to promote cell death. Our results suggest that DAPK3 acts as a pro-survival protein in melanoma and therefore is a promising, relatively novel drug target. As a kinase, it is highly druggable, and several inhibitors have already been developed, although they do not show specificity within the DAPK family [51]. The development of a more specific DAPK3 inhibitor would be warranted, since loss of expression and/or down-regulation of DAPK1 (and DAPK2) leads to tumorigenesis, and a wide acting inhibitor could potentially have contradictory effects on tumor growth and survival.
5. Conclusions
Our strategy to analyze the functional death signature of individual tumors has the potential to dramatically change the state-of-the-art of combinatorial drug therapy in precision cancer treatment, by reducing the number of persister cells surviving the initial treatment with minimal overlapping toxicities, thereby lessening the odds of developing drug resistance at later stages, and preventing tumor relapse. While we based our siRNA platform on V600E BRAF early passage melanoma cell lines in combination with vemurafenib, it can readily be adapted to any tumor from which primary cultures can be generated for running the siRNA platform together with standard targeted drug treatment. Furthermore, expansion of the screen to include proteins from additional death modules, such as ferroptosis and parthanatos [52], will further increase our understanding of the intact death pathways that can be targeted to achieve maximal tumor killing. Currently, we employed clinically approved drugs to target soft-spots; however, with additional drugs in the pipeline, as well as RNAi protocols expected to be available for clinical use in the near future, more soft-spots can be harnessed for personalized, combinatorial treatment of melanoma and other cancer.
Acknowledgments
We would like to thank Aya Shkedy, Lital Povodovski, Rivi Halimi, Maya David Teitelbaum, Liat Hammer, Yuval Gilad, Gal Chaim Nuta, and Avital Hay-Koren for providing significant assistance and crucial advice with experimental procedures and writing of this paper. This work was funded by a grant from the Israel Science Foundation, Grant No. 679/17 to AK.
Conflict of Interests
The authors declare no potential conflicts of interest or competing financial interests.
Supplementary Information
Please download the supplementary information from the below link
https://www.fortunejournals.com/supply/supp.JCSCT4547.pdf
References
- Wolf Y, Samuels Y. Cancer research in the era of immunogenomics. ESMO Open 3 (2018): 000475.
- Herrscher H, Robert C. Immune checkpoint inhibitors in melanoma in the metastatic, neoadjuvant, and adjuvant setting. Curr Opin Oncol 32 (2020): 106-113.
- Miller AJ, Mihm Jr MC. Melanoma. N Engl J Med (2006): 355:51-65.
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364 (2011): 2507-2516.
- Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 367 (2012): 1694-1703.
- Sullivan RJ, Infante JR, Janku F, et al. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: Results of a phase I dose-escalation and expansion study. Cancer Discov 8 (2018): 184-195.
- Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: Targeted strategies for melanoma. Nat Rev Cancer 12 (2012): 349-361.
- Sharma SV, Lee DY, Li B, et al. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell 141 (2010): 69-80.
- Hangauer MJ, Viswanathan VS, Ryan MJ, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551 (2017): 247-250.
- Shaffer SM, Dunagin MC, Torborg SR, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546 (2017): 431-435.
- Kochanowski K, Morinishi L, Altschuler SJ, et al. Drug persistence - From antibiotics to cancer therapies. Curr Opin Syst Biol 10 (2018): 1-8.
- Ramirez M, Rajaram S, Steininger RJ, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 7 (2016): 10690.
- Emery CM, Vijayendran KG, Zipser MC, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA 106 (2009): 20411–20416.
- Allen EM Van, Wagle N, Sucker A, et al. The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma. Cancer Discov 4 (2014): 94-109.
- Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480 (2011): 387-390.
- Ma XH, Piao SF, Dey S, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 124 (2014): 1406-1417.
- Locatelli SL, Careddu G, Stirparo GG, et al. Dual PI3K/ERK inhibition induces necroptotic cell death of Hodgkin Lymphoma cells through IER3 downregulation. Sci Rep 6 (2016): 35745.
- Cagnol S, Chambard J-CC. ERK and cell death: Mechanisms of ERK-induced cell death – apoptosis, autophagy and senescence. FEBS J 277 (2011): 2-21.
- Sanduja S, Feng Y, Mathis RA, Sokol ES, Reinhardt F, Halaban R, et al. AMPK promotes tolerance to Ras pathway inhibition by activating autophagy. Oncogene 35 (2016): 5295–303.
- Kinsey CG, Camolotto SA, Boespflug AM, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 25 (2019): 620-627.
- Rubinstein AD, Kimchi A. Life in the balance - A mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci 125 (2012): 5259-5268.
- Besser MJ, Shapira-Frommer R, Treves AJ, et al. Clinical responses in a phase II study using adoptive transfer of short-term cultured tumor infiltration lymphocytes in metastatic melanoma patients. Clin Cancer Res 16 (2010): 2646-2655.
- Cartlidge RA, Thomas GR, Cagnol S, et al. Oncogenic BRAFV600E inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res 21 (2008): 534-544.
- Massard C, Michiels S, Ferté C, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: Results of the MOSCATO 01 trial. Cancer Discov 7 (2017): 586-595.
- Flaherty KT, Gray R, Chen A, et al. The molecular analysis for therapy choice (NCI-MATCH) trial: lessons for genomic trial design. J Natl Cancer Inst 112 (2020): 1021-1029.
- Xu C, Nikolova O, Basom RS, et al. Functional precision medicine identifies novel druggable targets and therapeutic options in head and neck cancer. Clin Cancer Res 24 (2018): 2828-2843.
- Phan N, Hong JJ, Tofig B, et al. A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun Biol 2 (2019): 1-11.
- Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359 (2018): 920-926.
- Pauli C, Hopkins BD, Prandi D, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov 7 (2017): 462-477.
- Montero J, Sarosiek KA, Deangelo JD, et al. Drug-Induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 160 (2015): 977-989.
- Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA 105 (2008): 3041-3046.
- Sheridan C, Brumatti G, Martin SJ. Oncogenic B-RafV600E inhibits apoptosis and promotes ERK-dependent inactivation of Bad and Bim. J. Biol. Chem 283 (2019): 22128-22135.
- Lee EF, Harris TJ, Tran S, et al. BCL-XL and MCL-1 are the key BCL-2 family proteins in melanoma cell survival. Cell Death Dis 10 (2019): 342.
- Mckee CS, Hill DS, Redfern CPF, et al. Oncogenic BRAF signalling increases Mcl-1 expression in cutaneous metastatic melanoma. Exp Dermatol 22 (2013): 767-769.
- Mukherjee N, Lu Y, Almeida A, et al. Use of a MCL-1 inhibitor alone to de-bulk melanoma and in combination to kill melanoma initiating cells. Oncotarget 8 (2017): 46801-46817.
- Leverson JD, Phillips DC, Mitten MJ, et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci Transl Med 7 (2015): 279-279.
- Haq R, Yokoyama S, Hawryluk EB, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci USA 110 (2013): 4321-4326.
- Kotschy A, Szlavik Z, Murray J, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538 (2016): 477-482.
- Mulcahy Levy JM, Zahedi S, Griesinger AM, et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife 6 (2017): 19671.
- Bryant KL, Stalnecker CA, Zeitouni D, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 25 (2019): 628-640.
- Liu X, Wu J, Qin H, et al. The Role of Autophagy in the Resistance to BRAF Inhibition in BRAF-Mutated Melanoma. Target Oncol 13 (2018): 437-446.
- Li S, Song Y, Quach C, et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat Commun 10 (2019):1693.
- Ohashi Y, Tremel S, Williams RL. VPS34 complexes from a structural perspective. J Lipid Res 60 (2019): 229-241.
- Das TP, Suman S, Papu John AMS, et al. Activation of AKT negatively regulates the pro-apoptotic function of death-associated protein kinase 3 (DAPK3) in prostate cancer. Cancer Lett 377 (2016): 134-139.
- Kogel D, Reimertz C, Mech P, et al. Dlk/ZIP kinase-induced apoptosis in human medulloblastoma cells: requirement of the mitochondrial apoptosis pathway. Br J Cancer 85 (2001): 1801-1088.
- Tan J, Xianzhen J, Yin G, et al. Anacardic acid induces cell apoptosis of prostatic cancer through autophagy by ER stress/DAPK3/Akt signaling pathway. Oncol Rep 38 (2017): 1373-1382.
- Tang HW, Wang YB, Wang SL, et al. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J 30 (2011): 636-651.
- Kake S, Usui T, Ohama T, et al. Death-associated protein kinase 3 controls the tumor progression of A549 cells through ERK MAPK/c-Myc signaling. Oncol Rep 37 (2017): 1100-1106.
- Thomas F, Holmes KB, Kreuz S, et al. DAPK3 participates in the mRNA processing of immediate early genes in Chronic Lymphocytic Leukaemia. Mol Oncol. 2020;in press.
- Li J, Deng Z, Wang Z, et al. Zipper-interacting protein kinase promotes epithelial-mesenchymal transition, invasion and metastasis through AKT and NF-κB signaling and is associated with metastasis and poor prognosis in gastric cancer patients. Oncotarget 6 (2015): 8323-8338.
- Farag AK, Roh EJ. Death-associated protein kinase (DAPK) family modulators: Current and future therapeutic outcomes. Med Res Rev 39 (2019): 349-385.
- Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 25 (2018): 486-541.
